








生物技术通报 ›› 2025, Vol. 41 ›› Issue (3): 319-329.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0993
• 研究报告 • 上一篇
收稿日期:2024-10-12
出版日期:2025-03-26
发布日期:2025-03-20
通讯作者:
郭敏亮,男,博士,教授,研究方向 :根癌农杆菌与酶工程;E-mail: guoml@yzu.edu.cn作者简介:王浩,男,博士,讲师,研究方向 :微生物与酶工程;E-mail: wanghao@yzu.edu.cn
基金资助:
WANG Hao( ), CAO An-ni, GAO Xin-yi, GUO Min-liang(
), CAO An-ni, GAO Xin-yi, GUO Min-liang( )
)
Received:2024-10-12
Published:2025-03-26
Online:2025-03-20
摘要:
目的 原儿茶酸是一种应用广泛的酚类化合物,生物合成法生产原儿茶酸是替代高污染的化学合成法的潜在方案,寻找和改良合成原儿茶酸的酶是生物合成法的关键。根癌农杆菌甲氧基脱甲基酶Atu1420是一种能够将香草酸转化为原儿茶酸的酶,本研究旨在表征Atu1420的酶学特性,并对其进行改良。 方法 通过大肠杆菌表达了Atu1420,并利用高效液相色谱等方法测定了Atu1420的酶学特性。基于Atu1420的预测结构和催化机制,选择了19个位点对Atu1420进行了定点突变,利用4-氨基安替比林开发了一种可视化检测Atu1420催化活性的方法,用于筛选Atu1420定点突变的变异体。利用alphaFold对Atu1420变异体的结构进行预测,分析了变异体催化效率提升的潜在机制。 结果 确定了Atu1420的Vmax为33.5±1.6 nmol/(L·s),米氏常数Km为82.7±3.5 μmol/L,转换数kcat为(6.7±0.3)×10-1 s-1,kcat/Km为8.1×10-3 L/(μmol·s),最适pH在7-8之间,最适温度为30℃。本研究在定点突变的变异体中筛选出了5个酶活力增强的变异体,其中活力最强的变异体为G35S,其催化速度比野生型提高66.0%。对这5个位点进行组合突变,在酶活力方面并没有产生明显的叠加效应。结构的比对分析暗示121位精氨酸残基角度的偏转可能是造成变异体催化效率提升的原因。 结论 Atu1420的酶学特性得到了有效的表征,获得了5个催化效率提高的Atu1420变异体,变异体催化效率的提高可能是121位精氨酸残基角度的偏转造成的。
王浩, 曹安妮, 高欣怡, 郭敏亮. 根癌农杆菌甲氧基脱甲基酶Atu1420的酶学特性表征和定向进化[J]. 生物技术通报, 2025, 41(3): 319-329.
WANG Hao, CAO An-ni, GAO Xin-yi, GUO Min-liang. Enzymatic Characterization and Directed Evolution of Agrobacterium tumefaciens O-demethylase Atu1420[J]. Biotechnology Bulletin, 2025, 41(3): 319-329.

图1 Atu1420的表达和催化活性表征A:Atu1420催化的反应示意图;B:Atu1420的SDS-PAGE电泳图;C:HPLC检测Atu1420反应液峰图,蓝色曲线为不加Atu1420的反应液,红色曲线为加了Atu1420的反应液;D:Atu1420的酶动力学曲线,Vmax和Km通过双倒数作图法求出
Fig. 1 Identification of Atu1420 expression and catalytic activityA: Schematic diagram of the reaction catalyzed by Atu1420. B: SDS-PAGE electrophoretogram of Atu1420. C: Detecting peak of Atu1420 reaction mixture via HPLC. The blue curve inidicates the reaction mixture without Atu1420, and the red curve indicates the reaction mixture with Atu1420 added. D: Enzyme kinetics curve of Atu1420. The Vmax and Km values are obtained through the double reciprocal plot method
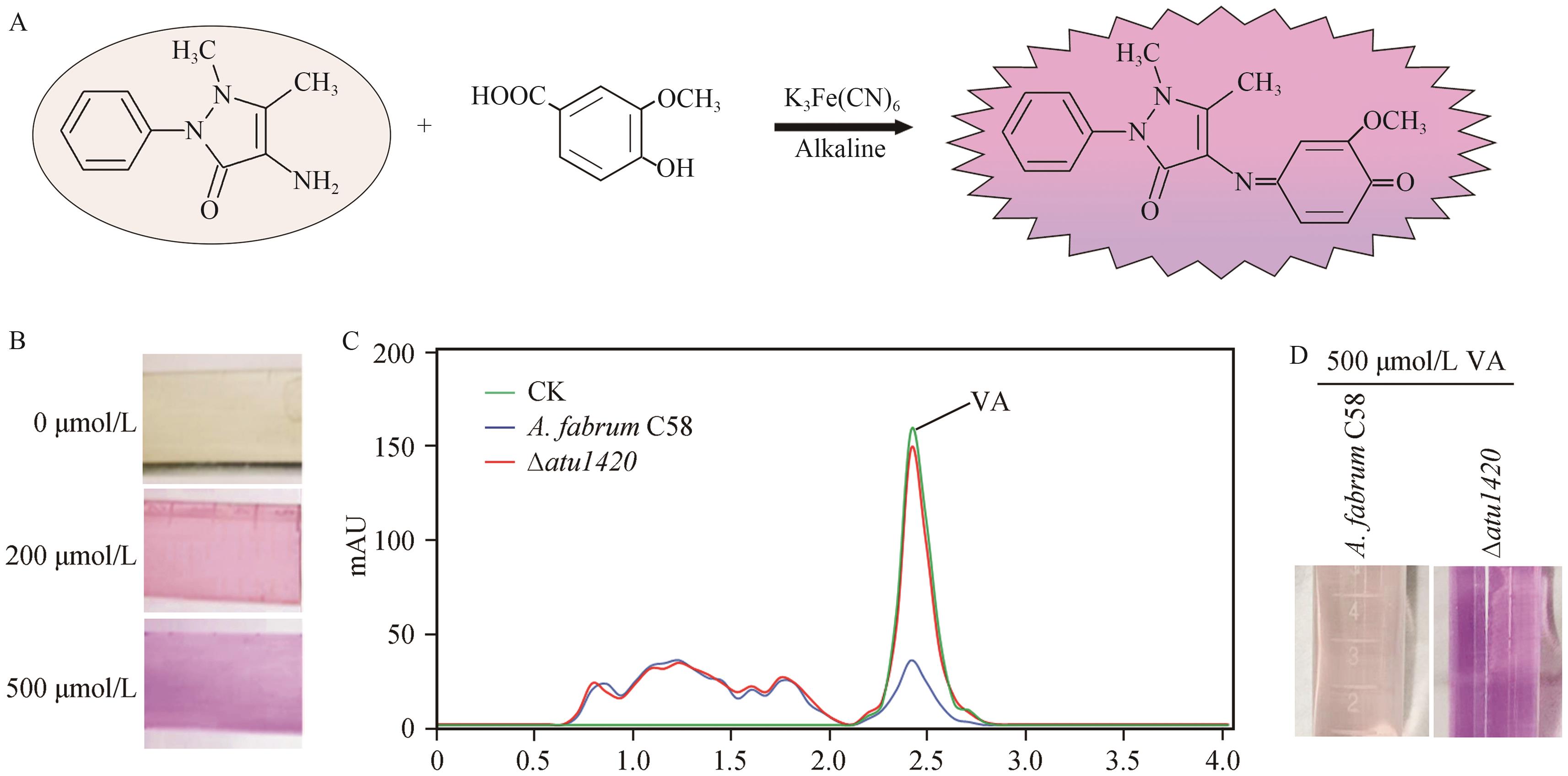
图3 4-氨基安替比林检测香草酸方法建立A:4-氨基安替比林与香草酸的颜色反应示意图; B:不同浓度香草酸与4-氨基安替比林反应的颜色差异; C:HPLC检测根癌农杆菌代谢香草酸的情况; CK为不加菌处理,其余均为加了根癌农杆菌处理6 h后香草酸的剩余情况; D:4-氨基安替比林法检测A. tumefaciens C58和∆atu1420分解香草酸的情况
Fig. 3 Establishment of the detection method for VA using 4-aminoantipyrineA: Schematic diagram of the reaction between 4-aminoantipyrine and VA. B: Colors in the reaction between 4-aminoantipyrine and VA of different concentrations. C: HPLC analysis of VA metabolism by A. tumefaciens. CK indicates the untreated control, while the other samples were treated with A. tumefaciens for 6 h. Curves shows the remaining VA levels. D: Detection of VA degradation by A. tumefaciens C58 and ∆atu1420 using the 4-aminoantipyrine method

图4 Atu1420的半理性设计和变异体的筛选A:香草酸与Atu1420的分子对接. 绿色分子为Atu1420,参与香草酸的结合和催化的4个氨基酸残基以棍棒形式呈现,蓝色表示N原子;靛蓝色分子为香草酸,红色表示氧原子。B:Atu1420同源蛋白的序列比对。1为来自Microbacterium trichothecenolyticum的Atu1420同源蛋白,2为来自Sinomonas atrocyanea的Atu1420同源蛋白,3为来自Citricoccus sp. K5的Atu1420同源蛋白,4为来自Sphingobium sp.的Atu1420同源蛋白,5为来自Rhizobium leguminosarum的Atu1420同源蛋白,6为来自Betaproteobacteria bacterium的Atu1420同源蛋白,7为Atu1420,蓝色三角箭头指示的是A图中4个关键氨基酸残基,紫色星号指示的是与这4个关键氨基酸在空间上邻近的氨基酸残基,数字指示的是在Atu1420中的氨基酸位点。C:菌水平检测Atu1420及其变异体的催化活性。CK为不加菌处理,其余均为加了根癌农杆菌处理6 h后香草酸的剩余情况。D:蛋白质水平检测Atu1420及其变异体的催化活性。CK为不加蛋白质的处理,其余均为加了蛋白质处理30 min后原儿茶酸的生成情况。E:菌水平检测多点组合突变变异体的催化活性。CK为不加菌处理,其余均为加了根癌农杆菌处理6 h后香草酸的剩余情况。F:蛋白质水平检测多点组合突变变异体的催化活性。CK为不加蛋白质的处理,其余均为加了蛋白质处理30 min后原儿茶酸的生成情况
Fig. 4 Semi-rational design and screening of variants of Atu1420A: Molecular docking of VA with Atu1420. The green molecule refers to Atu1420, with four amino acid residues involved in VA binding and catalysis shown in stick form, where blue indicates nitrogen atoms. The cyan molecule indicates VA, and red indicates oxygen atoms. B: Sequence alignment of Atu1420 homologous proteins. 1 is the Atu1420 homologous protein from Microbacterium trichothecenolyticum, 2 from Sinomonas atrocyanea, 3 from Citricoccus sp. K5, 4 from Sphingobium sp., 5 from Rhizobium leguminosarum, 6 from Betaproteobacteria bacterium, and 7 is Atu1420 itself. Blue triangular arrows indicate the four key amino acid residues from Fig. A, while purple asterisks indicate amino acid residues spatially adjacent to these four key residues. Numbers indicate amino acid positions in Atu1420. C: Catalytic activity of Atu1420 and its variants in the bacteria. CK indicates the control without adding A. tumefaciens, while the other samples were treated with A. tumefaciens for 6 h. D: Catalytic activity of Atu1420 and its variants using proteins. CK indicates the control without adding protein, while the other samples were treated with the protein for 30 min. E: Catalytic activity of multi-points combined variants in the bacteria. CK indicates the control without bacteria, while the other samples were treated with A. tumefaciens for 6 h. F: Catalytic activity of multi-points combined variants using proteins. CK indicates the control without adding protein, while the other samples were treated with the protein for 30 min

图6 Atu1420点突变变异体的结构分析绿色碳骨架的基团为Atu1420的4个关键氨基酸残基,其他颜色碳骨架的基团为各点突变体的4个关键氨基酸残基,靛蓝色碳骨架的分子为香草酸
Fig. 6 Structural analysis of Atu1420 mutated variantsThe green carbon skeleton indicate the four key amino acid residues of Atu1420, and the carbon skeletons with other colors indicate the four key amino acid residues of other variants. The cyan carbon skeleton molecule indicates VA
| 1 | Mahfuz S, Mun HS, Dilawar MA, et al. Potential role of protocatechuic acid as natural feed additives in farm animal production [J]. Animals, 2022, 12(6): 741. |
| 2 | Kakkar S, Bais S. A review on protocatechuic acid and its pharmacological potential [J]. ISRN Pharmacol, 2014, 2014: 952943. |
| 3 | Masella R, Santangelo C, D'Archivio M, et al. Protocatechuic acid and human disease prevention: biological activities and molecular mechanisms [J]. Curr Med Chem, 2012, 19(18): 2901-2917. |
| 4 | Market and Research 2020. Global Protocatechuic Acid (CAS99-50-3) Market 2020 by Manufacturers, Regions, Type and Application, Forecast to 2025 [R]. Pune: Marketsandresearch.biz, p.156. |
| 5 | Li J, Fu JL, Yue C, et al. Highly efficient biosynthesis of protocatechuic acid via recombinant Pseudomonas putida KT2440 [J]. J Agric Food Chem, 2023, 71(27): 10375-10382. |
| 6 | Quan W, Xu Y, Xie YT, et al. In vitro antioxidant properties and phenolic profile of acid aqueous ethanol extracts from Torreya grandis seed coat [J]. Molecules, 2022, 27(17): 5560. |
| 7 | Antony FM, Wasewar K. Reactive extraction: a promising approach to separate protocatechuic acid [J]. Environ Sci Pollut Res Int, 2020, 27(22): 27345-27357. |
| 8 | 邱笛, 周超, 张根林. 工程微生物合成香草醛的进展与挑战 [J]. 生物加工过程, 2023, 21(4): 355-367. |
| Qiu D, Zhou C, Zhang GL. Advances and challenges in the synthesis of vanillin by engineered microorganisms [J]. Chin J Bioprocess Eng, 2023, 21(4): 355-367. | |
| 9 | 刘超, 邓裕斌, 武书彬. 木质素制备香兰素的方法对比分析 [J]. 造纸科学与技术, 2014, 33(6): 53-57, 99. |
| Liu C, Deng YB, Wu SB. Comparative analysis of vanillin preparation methods from lignin [J]. Pap Sci Technol, 2014, 33(6): 53-57, 99. | |
| 10 | 王鑫鑫, 徐宇骋, 刘翠翠, 等. 一种生物化学法制备原儿茶酸的方法. CN202010921942.9 [P]. 2020-09-04. |
| Wang XX, Xu YC, Liu CC, et al. A method for preparing protocatechuic acid by biochemical method. CN202010921942.9 [P]. 2020-09-04. | |
| 11 | Li K, Frost JW. Synthesis of vanillin from glucose [J]. J Am Chem Soc, 1998, 120(40): 10545-10546. |
| 12 | Wang M, Wang HM, Gao C, et al. Efficient production of protocatechuic acid using systems engineering of Escherichia coli [J]. Metab Eng, 2024, 82: 134-146. |
| 13 | Moriwaki Y, Yato M, Terada T, et al. Understanding the molecular mechanism underlying the high catalytic activity of p-hydroxybenzoate hydroxylase mutants for producing Gallic acid [J]. Biochemistry, 2019, 58(45): 4543-4558. |
| 14 | Upadhyay P, Lali A. Protocatechuic acid production from lignin-associated phenolics [J]. Prep Biochem Biotechnol, 2021, 51(10): 979-984. |
| 15 | Harada A, Kamimura N, Takeuchi K, et al. The crystal structure of a new O-demethylase from Sphingobium sp. strain SYK-6 [J]. FEBS J, 2017, 284(12): 1855-1867. |
| 16 | Rosini E, D'Arrigo P, Pollegioni L. Demethylation of vanillic acid by recombinant LigM in a one-pot cofactor regeneration system [J]. Catal Sci Technol, 2016, 6(21): 7729-7737. |
| 17 | Bleem AC, Kuatsjah E, Johnsen J, et al. Evolution and engineering of pathways for aromatic O-demethylation in Pseudomonas putida KT2440 [J]. Metab Eng, 2024, 84: 145-157. |
| 18 | Bhattacharya A, Sood P, Citovsky V. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection [J]. Mol Plant Pathol, 2010, 11(5): 705-719. |
| 19 | Lassalle F, Campillo T, Vial L, et al. Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens [J]. Genome Biol Evol, 2011, 3: 762-781. |
| 20 | Wang H, Zhang MQ, Wang EY, et al. Agrobacterium fabrum gene atu1420 regulates the pathogenicity by affecting the degradation of growth- and virulence-associated phenols [J]. Res Microbiol, 2023, 174(3): 104011. |
| 21 | Sambrook J, Fristch EF, Manlatis T. Molecular Cloning: A Laboratory Manual [M]. 2nd ed. New York: Cold Spring Harbor Laboratory Press, 1989: 20-25. |
| 22 | Cangelosi GA, Best EA, Martinetti G, et al. Genetic analysis of Agrobacterium [J]. Methods Enzymol, 1991, 204: 384-397. |
| 23 | Gelvin SB. Agrobacterium virulence gene induction [J]. Methods Mol Biol, 2006, 343: 77-84. |
| 24 | Wang H, Zhang MQ, Xu YJ, et al. Agrobacterium fabrum atu0526-encoding protein is the only chemoreceptor that regulates chemoattraction toward the broad antibacterial agent formic acid [J]. Biology, 2021, 10(12): 1345. |
| 25 | Chen H, Huang MF, Yan WL, et al. Enzymatic regio- and enantioselective C-H oxyfunctionalization of fatty acids [J]. ACS Catal, 2021, 11(16): 10625-10630. |
| 26 | Mosae Selvakumar P. Phenol sensing studies by 4-aminoantipyrine method-a review [J]. Org Med Chem Int J, 2018, 5(2): 555657. |
| 27 | Parke D. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens [J]. J Bacteriol, 1995, 177(13): 3808-3817. |
| 28 | Kohler AC, Mills MJL, Adams PD, et al. Structure of aryl O-demethylase offers molecular insight into a catalytic tyrosine-dependent mechanism [J]. Proc Natl Acad Sci USA, 2017, 114(16): E3205-E3214. |
| 29 | Chica RA, Doucet N, Pelletier JN. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design [J]. Curr Opin Biotechnol, 2005, 16(4): 378-384. |
| 30 | Senior AW, Evans R, Jumper J, et al. Improved protein structure prediction using potentials from deep learning [J]. Nature, 2020, 577(7792): 706-710. |
| 31 | Reetz M. Making enzymes suitable for organic chemistry by rational protein design [J]. Chembiochem, 2022, 23(14): e202200049. |
| 32 | Savile CK, Janey JM, Mundorff EC, et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture [J]. Science, 2010, 329(5989): 305-309. |
| 33 | Novick SJ, Dellas N, Garcia R, et al. Engineering an amine transaminase for the efficient production of a chiral sacubitril precursor [J]. ACS Catal, 2021, 11(6): 3762-3770. |
| 34 | Xie YY, Xu F, Yang L, et al. Engineering the large pocket of an (S)-selective transaminase for asymmetric synthesis of (S)-1-amino-1-phenylpropane [J]. Catal Sci Technol, 2021, 11(7): 2461-2470. |
| [1] | 陆峰, 黄玉红, 林燕娜, 马富强. CO2还原用甲酸脱氢酶分子改造的研究进展[J]. 生物技术通报, 2025, 41(3): 14-24. |
| [2] | 乔烨, 张楠, 杨建花, 张翠英, 朱蕾蕾. 糖磷酸酶的挖掘及其酶学性质研究[J]. 生物技术通报, 2024, 40(7): 299-306. |
| [3] | 杨秉乾, 恽辰珂, 常思源, 郭盛, 张森. 丹参药渣木质素降解菌的分离及酶学特性[J]. 生物技术通报, 2024, 40(11): 269-276. |
| [4] | 张开平, 刘燕丽, 涂绵亮, 李继伟, 吴文标. 烟曲霉A-16产纤维素酶工艺优化及酶学特性[J]. 生物技术通报, 2022, 38(9): 215-225. |
| [5] | 付巧, 林啟兰, 薛强, 熊海容, 王亚伟. N端截短CBM41对枯草芽孢杆菌来源普鲁兰酶酶学性质的影响[J]. 生物技术通报, 2022, 38(6): 245-251. |
| [6] | 张雪, 谭玉萌, 蒋海霞, 杨广宇. 基于单细胞超高通量筛选的α-1,2-岩藻糖基转移酶定向进化[J]. 生物技术通报, 2022, 38(1): 289-298. |
| [7] | 郝俊尧, 马富强, 杨广宇. 产碱杆菌Alcaligenes sp.KS-85来源肌酸酶活性中心的关键氨基酸功能研究[J]. 生物技术通报, 2021, 37(3): 75-83. |
| [8] | 邱锦, 黄火清, 姚斌, 罗会颖. 解淀粉芽孢杆菌淀粉酶催化活力改良及其在枯草芽孢杆菌中的高效表达[J]. 生物技术通报, 2019, 35(9): 134-143. |
| [9] | 郭静静, 郭磊磊, 赵云岫, 戴亦军. Ensifer meliloti 1021烟酰胺酶的酶学特性及3-氰基吡啶调控机理的研究[J]. 生物技术通报, 2019, 35(8): 51-58. |
| [10] | 秦日甜, 谢占玲. 镰刀菌Q7-31T果胶酶PGL1的分离纯化、酶学性质鉴定及结构分析[J]. 生物技术通报, 2018, 34(4): 151-160. |
| [11] | 任天雷, 杨海泉, 许菲. 基于分子结构与生物信息学等多维度特征的定向进化改造甲基对硫磷水解酶[J]. 生物技术通报, 2018, 34(10): 194-200. |
| [12] | 于林港, 宿玲恰, 吴敬. Escherichia coli str. K-12 substr. MG1655海藻糖酶Tre F的重组表达及性质研究[J]. 生物技术通报, 2017, 33(4): 177-184. |
| [13] | 贾博涵,周伟,赵罗迪,杨埔,苟敏. 一株产纤维素酶细菌的分离鉴定及酶学特性研究[J]. 生物技术通报, 2014, 30(11): 187-192. |
| [14] | 张丽靖;沈江锋;金庆超;杨郁;. 一株酸性淀粉酶产生菌的分离、鉴定及酶学特性初步研究[J]. , 2011, 0(05): 142-145. |
| [15] | 谢振荣;慕跃林;闫丽娟;赵春雷;黄遵锡;. α-葡萄糖苷酶高产菌株HB-9-5的选育及产酶条件的优化[J]. , 2010, 0(06): 206-211. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||