








生物技术通报 ›› 2025, Vol. 41 ›› Issue (5): 70-81.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1104
王轶民( ), 李莹, 董海涛, 张恒瑞, 常璐, 高田甜, 韩德俊(
), 李莹, 董海涛, 张恒瑞, 常璐, 高田甜, 韩德俊( ), 吴建辉(
), 吴建辉( )
)
收稿日期:2024-11-14
出版日期:2025-05-26
发布日期:2025-06-05
通讯作者:
吴建辉,男,博士,副教授,研究方向 :抗病遗传与分子育种;E-mail: wujh@nwafu.edu.cn作者简介:王轶民,女,硕士研究生,研究方向 :作物分子生物学基础;E-mail: wangyimin2022@nwafu.edu.cn基金资助:
WANG Yi-min( ), LI Ying, DONG Hai-tao, ZHANG Heng-rui, CHANG Lu, GAO Tian-tian, HAN De-jun(
), LI Ying, DONG Hai-tao, ZHANG Heng-rui, CHANG Lu, GAO Tian-tian, HAN De-jun( ), WU Jian-hui(
), WU Jian-hui( )
)
Received:2024-11-14
Published:2025-05-26
Online:2025-06-05
摘要:
目的 旨在系统鉴定和分析小麦SRO基因家族的结构特征、进化规律及其在生物胁迫条件下的表达模式,揭示其在小麦胁迫响应中的潜在作用。 方法 以拟南芥(Arabidopsis thaliana)、番茄(Solanum lycopersicum)和玉米(Zea mays)为参考,对14个小麦族物种的36个基因组进行SRO基因家族鉴定,并对小麦属中的二倍体(Triticum urartu v2.0)、四倍体(Wild Emmer v1.0)以及六倍体(Chinese Spring v2.1)基因组中鉴定到的SRO家族成员进行理化性质、保守基序、基因结构、系统发育、顺式调控元件、共线性和蛋白质-蛋白质相互作用网络分析。 结果 蛋白质理化性质分析结果表明,SRO家族成员蛋白普遍偏碱性且具有亲水性;基因结构分析显示,所有成员均存在内含子,且序列长度差异大;系统发育分析表明,小麦SRO和玉米SRO具有更近的进化关系;启动子顺式调控元件分析发现,SRO家族成员主要包含响应植物生物和非生物胁迫相关的元件;进化选择和核苷酸多样性分析表明,小麦SRO家族成员从二倍体形成四倍体的过程中受到了较强的正向选择压力,而在形成六倍体后趋于稳定;在小麦条锈菌胁迫下,六倍体小麦SRO家族成员呈现不同的表达模式。 结论 小麦SRO基因家族在二倍体向四倍体的进化过程中经历了强烈的正向选择,并在六倍体小麦中趋于稳定。部分成员在生物胁迫条件下与对照相比表现出显著的表达差异。
王轶民, 李莹, 董海涛, 张恒瑞, 常璐, 高田甜, 韩德俊, 吴建辉. SRO家族蛋白在小麦多倍化进程中的演化规律[J]. 生物技术通报, 2025, 41(5): 70-81.
WANG Yi-min, LI Ying, DONG Hai-tao, ZHANG Heng-rui, CHANG Lu, GAO Tian-tian, HAN De-jun, WU Jian-hui. Evolutionary Patterns of SRO Family Proteins in the Polyploidization Process of Wheat[J]. Biotechnology Bulletin, 2025, 41(5): 70-81.
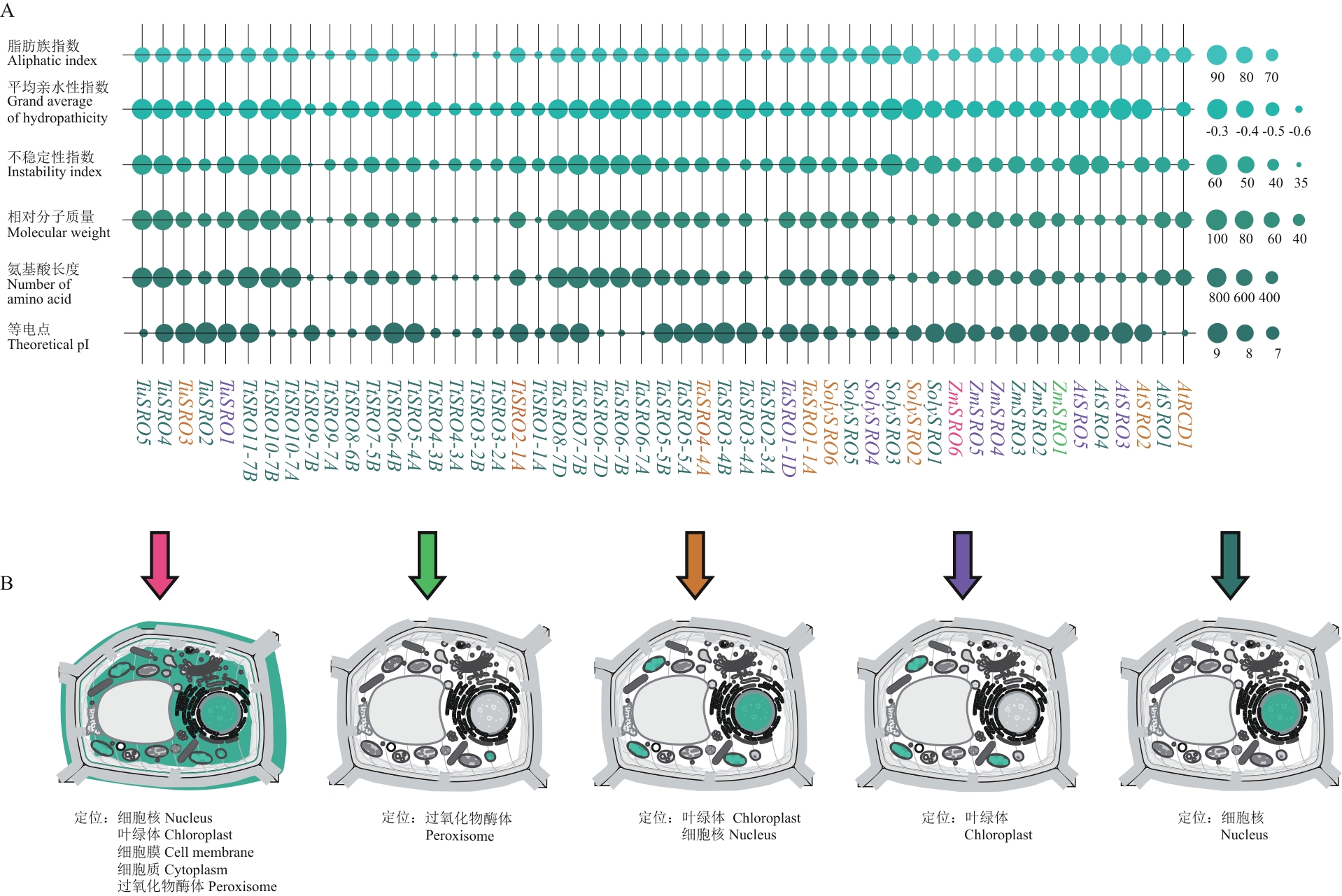
图1 SRO基因家族成员理化性质以及亚细胞定位预测A:SRO基因家族成员理化性质分析(At:拟南芥;Zm:玉米;Soly:番茄;Ta:六倍体小麦;Tt:四倍体小麦;Tu:二倍体小麦,下同);B:SRO基因家族成员亚细胞定位预测(箭头的颜色与图A中SRO家族成员名字颜色对应)
Fig. 1 Physicochemical properties and subcellular localization prediction of SRO gene family membersA: Physiochemical properties analysis of SRO gene family members (At: Arabidopsis thaliana; Zm: Zea mays; Soly: Solanum lycopersicum; Ta: hexaploid wheat; Tt: tetraploid wheat; Tu: diploid wheat. The same below). B: Subcellular localization prediction of SRO gene family members (The color of the arrows corresponds to the colors of the SRO family member names in Fig. A)

图2 SRO基因家族进化树、保守基序分析、基因结构和结构域分析A:SRO基因家族进化树;B:SRO基因家族保守基序分析C:SRO基因家族基因结构分析;D:SRO基因家族结构域分析
Fig. 2 Phylogenetic tree, conserved motifs analysis, and gene structure and domain analysis of SRO gene familyA: Phylogenetic tree of the SRO gene family. B: Conserved motif analysis of the SRO gene family. C: Gene structure analysis of the SRO gene family. D: Domain analysis of the SRO gene family
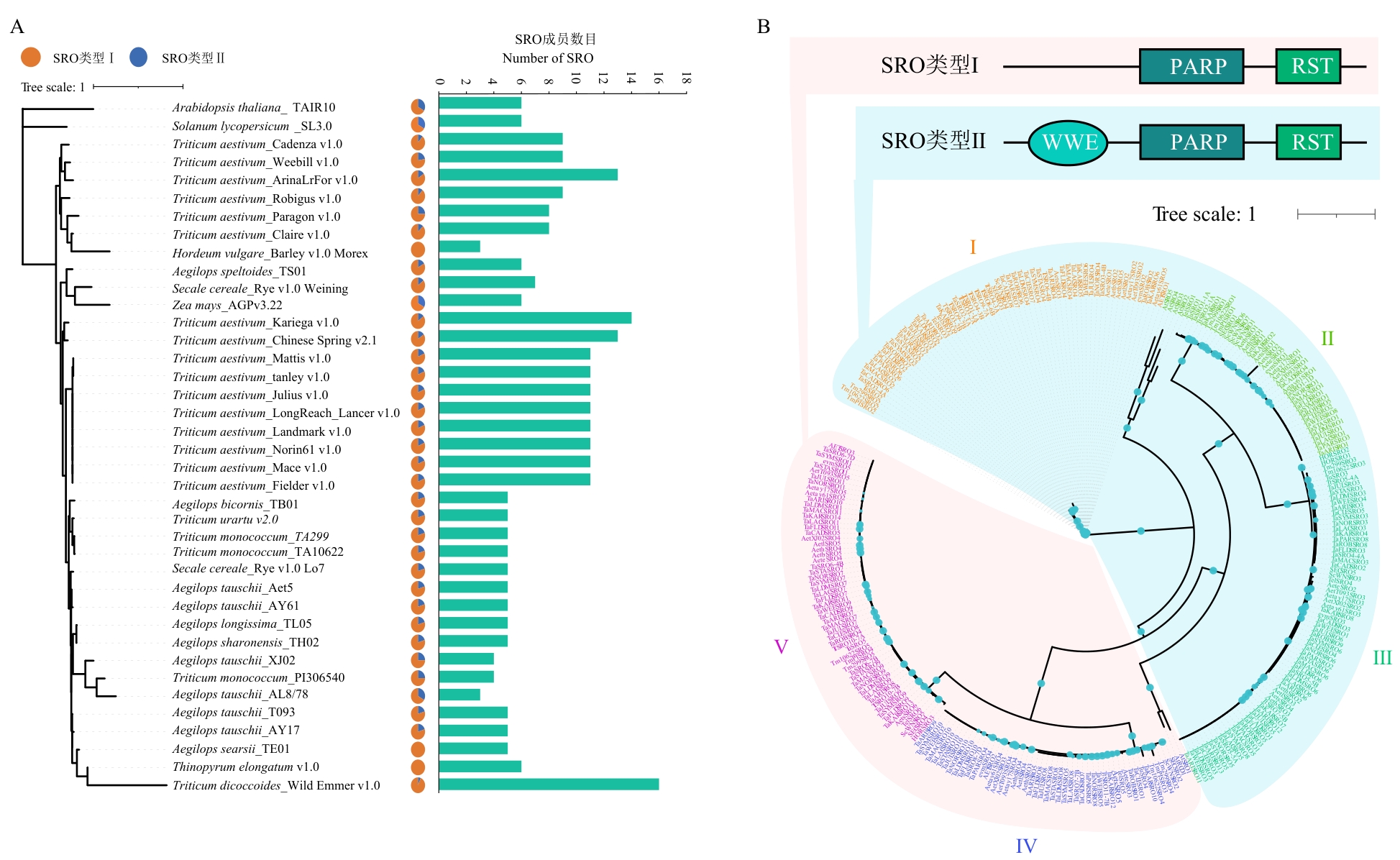
图3 系统发育分析A:麦族系统发育进化树以及各物种SRO成员数目;B:SRO家族成员物种进化树
Fig. 3 Phylogenetic analysisA: Phylogenetic tree of the Triticeae tribe and the number of SRO members in each species. B: Species phylogenetic tree of SRO family members

图4 启动子顺式调控元件分析A:小麦二/四/六倍体SRO家族成员响应调控元件的具体数量;B:小麦二/四/六倍体SRO家族成员响应三类调控元件的总数
Fig. 4 Analysis of promoter cis-regulatory elementsA: The specific number of responsive regulatory elements for SRO family members in diploid, tetraploid, and hexaploid wheat. B: The total number of responsive regulatory elements of three types for SRO family members in diploid, tetraploid, and hexaploid wheat

图5 SRO家族成员进化选择分析和TuSRO、TtSRO和TaSRO之间的多序列比对A:核苷酸多样性以及Ka/Ks分析;B:TuSRO、TtSRO和TaSRO之间的多序列比对
Fig. 5 Evolutionary selection analysis of the SRO family members and multiple sequence alignment among TuSRO, TtSRO, and TaSROA: Nucleotide diversity and Ka/Ks analysis. B: The sequence alignment between TuSRO, TtSRO, and TaSRO

图7 TaSRO群体表达模式分析及互作预测分析A:TaSRO群体表达量分析;B:TaSRO1-1A互作预测分析
Fig. 7 Expression pattern analysis and interaction prediction of TaSRO gene familyA: Expression analysis of TaSRO gene family. B: Interaction prediction analysis of TaSRO1-1A
| 1 | Savary S, Willocquet L, Pethybridge SJ, et al. The global burden of pathogens and pests on major food crops [J]. Nat Ecol Evol, 2019, 3(3): 430-439. |
| 2 | Zeng QD, Zhao J, Wu JH, et al. Wheat stripe rust and integration of sustainable control strategies in China [J]. Front Agr Sci Eng, 2022, 9(1): 37. |
| 3 | 陈万权, 康振生, 马占鸿, 等. 中国小麦条锈病综合治理理论与实践 [J]. 中国农业科学, 2013, 46(20): 4254-4262. |
| Chen WQ, Kang ZS, Ma ZH, et al. Integrated Management of Wheat Stripe Rust Caused by Puccinia striiformis f.sp. tritici in China [J]. Sci Agric Sin, 2013, 46(20): 4254-4262. | |
| 4 | Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal [J]. Genes Dev, 2005, 19(17): 1951-1967. |
| 5 | Schreiber V, Dantzer F, Ame JC, et al. Poly(ADP-ribose): novel functions for an old molecule [J]. Nat Rev Mol Cell Biol, 2006, 7(7): 517-528. |
| 6 | Bai P. Biology of poly(ADP-ribose) polymerases: the factotums of cell maintenance [J]. Mol Cell, 2015, 58(6): 947-958. |
| 7 | Perina D, Mikoč A, Ahel J, et al. Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life [J]. DNA Repair, 2014, 23: 4-16. |
| 8 | Kong L, Feng BM, Yan Y, et al. Noncanonical mono(ADP-ribosyl)ation of zinc finger SZF proteins counteracts ubiquitination for protein homeostasis in plant immunity [J]. Mol Cell, 2021, 81(22): 4591-4604.e8. |
| 9 | Gao XQ, Gao GG, Zheng WF, et al. PARylation of 14-3-3 proteins controls the virulence of Magnaporthe oryzae [J]. Nat Commun, 2024, 15(1): 8047. |
| 10 | Jaspers P, Overmyer K, Wrzaczek M, et al. The RST and PARP-like domain containing SRO protein family: analysis of protein structure, function and conservation in land plants [J]. BMC Genomics, 2010, 11: 170. |
| 11 | Ahlfors R, Lång S, Overmyer K, et al. Arabidopsis radical-induced cell death1 belongs to the wwe protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses [J]. Plant Cell, 2004, 16(7): 1925-1937. |
| 12 | You J, Zong W, Du H, et al. A special member of the rice SRO family, OsSRO1c, mediates responses to multiple abiotic stresses through interaction with various transcription factors [J]. Plant Mol Biol, 2014, 84(6): 693-705. |
| 13 | Jiang HH, Xiao Y, Zhu SW. Genome-wide identification, systematic analysis and characterization of SRO family genes in maize (Zea mays L.) [J]. Acta Physiol Plant, 2018, 40(10): 176. |
| 14 | 李保珠, 赵翔, 赵孝亮, 等. 拟南芥SRO蛋白家族的结构及功能分析 [J]. 遗传, 2013, 35(10): 1189-1197. |
| Li BZ, Zhao X, Zhao XL, et al. Structure and function analysis of Arabidopsis thaliana SRO protein family [J]. Hereditas, 2013, 35(10): 1189-1197. | |
| 15 | Liu ST, Liu SW, Wang M, et al. A wheat SIMILAR TO RCD-ONEgene enhances seedling growth and abiotic stress resistance by modulating redox homeostasis and maintaining genomic integrity [J]. The Plant Cell, 2014,26(1): 164-180. |
| 16 | Qin LM, Kong FF, Wei L, et al. Maize ZmSRO1e promotes mesocotyl elongation and deep sowing tolerance by inhibiting the activity of ZmbZIP61 [J]. J Integr Plant Biol, 2024, 66(8): 1571-1586. |
| 17 | Teotia S, Lamb RS. The paralogous genes radical-induced cell death1 and similar to rcd one1 have partially redundant functions during Arabidopsis development [J]. Plant Physiol, 2009, 151(1): 180-198. |
| 18 | Teotia S, Lamb RS. RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana [J]. J Exp Bot, 2011, 62(3): 1271-1284. |
| 19 | 赵秋芳, 马海洋, 贾利强, 等. 玉米SRO基因家族的鉴定及表达分析 [J]. 中国农业科学, 2018, 51(15): 196-206. |
| Zhao QF, Ma HY, Jia LQ, et al. Genome-wide identification and expression analysis of SRO genes family in maize [J]. Sci Agric Sin, 2018, 51(15): 196-206. | |
| 20 | Gao HJ, Cui JJ, Liu SX, et al. Natural variations of ZmSRO1d modulate the trade-off between drought resistance and yield by affecting ZmRBOHC-mediated stomatal ROS production in maize [J]. Mol Plant, 2022, 15(10): 1558-1574. |
| 21 | Qiao YL, Gao XQ, Liu ZC, et al. Genome-wide identification and analysis of SRO gene family in Chinese cabbage (Brassica rapa L.) [J]. Plants, 2020, 9(9): 1235. |
| 22 | Li N, Xu RQ, Wang BK, et al. Genome-wide identification and evolutionary analysis of the SRO gene family in tomato [J]. Front Genet, 2021, 12: 753638. |
| 23 | Wang M, Wang M, Zhao M, et al. TaSRO1 plays a dual role in suppressing TaSIP1 to fine tune mitochondrial retrograde signalling and enhance salinity stress tolerance [J]. New Phytol, 2022, 236(2): 495-511. |
| 24 | Liu SP, Li L, Wang WL, et al. TaSRO1 interacts with TaVP1 to modulate seed dormancy and pre-harvest sprouting resistance in wheat [J]. J Integr Plant Biol, 2024, 66(1): 36-53. |
| 25 | Ma SW, Wang M, Wu JH, et al. WheatOmics: a platform combining multiple omics data to accelerate functional genomics studies in wheat [J]. Mol Plant, 2021, 14(12): 1965-1968. |
| 26 | Mistry J, Chuguransky S, Williams L, et al. Pfam: the protein families database in 2021 [J]. Nucleic Acids Res, 2021, 49(D1): D412-D419. |
| 27 | Letunic I, Khedkar S, Bork P. SMART: recent updates, new developments and status in 2020 [J]. Nucleic Acids Res, 2021, 49(D1): D458-D460. |
| 28 | Horton P, Park KJ, Obayashi T, et al. WoLF PSORT: protein localization predictor [J]. Nucleic Acids Res, 2007, 35(Web Server issue): W585-W587. |
| 29 | Proost S, Mutwil M. CoNekT: an open-source framework for comparative genomic and transcriptomic network analyses [J]. Nucleic Acids Res, 2018, 46(W1): W133-W140. |
| 30 | Portwood JL 2nd, Woodhouse MR, Cannon EK, et al. MaizeGDB 2018: the maize multi-genome genetics and genomics database [J]. Nucleic Acids Res, 2019, 47(D1): D1146-D1154. |
| 31 | 郭安源, 朱其慧, 陈新, 等. GSDS: 基因结构显示系统 [J]. 遗传, 2007, 29(8): 1023-1026. |
| Guo AY, Zhu QH, Chen X, et al. GSDS a gene structure display server [J]. Hereditas, 2007, 29(8): 1023-1026. | |
| 32 | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching [J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| 33 | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| 34 | Letunic I, Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation [J]. Bioinformatics, 2007, 23(1): 127-128. |
| 35 | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| 36 | Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets [J]. Mol Biol Evol, 2017, 34(12): 3299-3302. |
| 37 | Wang DP, Zhang YB, Zhang Z, et al. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies [J]. Genomics Proteomics Bioinformatics, 2010, 8(1): 77-80. |
| 38 | Schwede T, Kopp J, Guex N, et al. SWISS-MODEL: an automated protein homology-modeling server [J]. Nucleic Acids Res, 2003, 31(13): 3381-3385. |
| 39 | Lopes I, Altab G, Raina P, et al. Gene size matters: an analysis of gene length in the human genome [J]. Front Genet, 2021, 12: 559998. |
| 40 | Qin LM, Sun L, Wei L, et al. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress [J]. Plant J, 2021, 105(4): 1010-1025. |
| 41 | 张小莉, 赵孝亮, 李保珠, 等. 拟南芥SRO1调节植物重金属汞胁迫应答 [J]. 科学通报, 2014, 59(20): 1967-1974. |
| Zhang XL, Zhao XL, Li BZ, et al. Arabidopsis SRO1 regulates the response of heavy metal mercury stress in plants [J]. Chin Sci Bull, 2014, 59(20): 1967-1974. | |
| 42 | Zhao XL, Gao LJ, Jin PN, et al. The similar to RCD-one 1 protein SRO1 interacts with GPX3 and functions in plant tolerance of mercury stress [J]. Biosci Biotechnol Biochem, 2018, 82(1): 74-80. |
| 43 | Yang YQ, Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses [J]. New Phytol, 2018, 217(2): 523-539. |
| 44 | Munns R, Gilliham M. Salinity tolerance of crops-what is the cost? [J]. New Phytol, 2015, 208(3): 668-673. |
| 45 | Borsani O, Zhu JH, Verslues PE, et al. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis [J]. Cell, 2005, 123(7): 1279-1291. |
| 46 | Hernandez-Garcia CM, Finer JJ. Identification and validation of promoters and cis-acting regulatory elements [J]. Plant Sci, 2014, 217/218: 109-119. |
| 47 | 胡雅丹, 伍国强, 刘晨, 等. MYB转录因子在调控植物响应逆境胁迫中的作用 [J]. 生物技术通报, 2024, 40(6): 5-22. |
| Hu YD, Wu GQ, Liu C, et al. The role of MYB transcription factor in regulating plant response to adversity stress [J]. Biotechnol Bull, 2024, 40(6): 5-22 . | |
| 48 | 谢鹏飞, 朱蕾, 冯玲, 等. 转录因子MYC2介导植物抗生物胁迫的研究进展 [J]. 应用昆虫学报, 2020, 57(4): 781-787. |
| Xie PF, Zhu L, Feng L, et al. Research progress in transcription factor MYC2 mediating plant resistance to biological stress [J]. Chin J Appl Entomol, 2020, 57(4): 781-787. | |
| 49 | Li W, Deng YW, Ning YS, et al. Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding [J]. Annu Rev Plant Biol, 2020, 71: 575-603. |
| 50 | 刘锐涛, 张颖, 樊秀彩, 等. 一氧化氮在植物抗病反应中的作用机制 [J]. 植物生理学报, 2020, 56(4): 625-634. |
| Liu RT, Zhang Y, Fan XC, et al. Mechanism of nitric oxide in plant response and resistance to disease [J]. Plant Physiol J, 2020, 56(4): 625-634. | |
| 51 | 王立超, 李欢, 盛若成, 等. 乙酰化修饰在植物病原物致病过程中的作用 [J]. 生物技术通报, 2024, 40(5): 1-12. |
| Wang LC, Li H, Sheng RC, et al. Role of acetylation in the pathogenic process of plant pathogens [J]. Biotechnol Bull, 2024, 40(5): 1-12. |
| [1] | 王伟伟, 赵振杰, 王志, 邹景伟, 罗政辉, 张玉杰, 钮力亚, 于亮, 杨学举. 盐胁迫下与小麦生理响应相关的耐盐基因研究进展[J]. 生物技术通报, 2025, 41(5): 14-22. |
| [2] | 王田田, 常雪瑞, 黄婉洋, 黄嘉欣, 苗如意, 梁燕平, 王静. 辣椒GASA基因家族的鉴定及分析[J]. 生物技术通报, 2025, 41(4): 166-175. |
| [3] | 黄金恒, 黄茜, 张家燕, 周新裕, 廖沛然, 杨全. 广金钱草C3H基因家族鉴定及不同品种表达分析[J]. 生物技术通报, 2025, 41(4): 243-255. |
| [4] | 马耀武, 张麒宇, 杨淼, 蒋诚, 张振宇, 张伊琳, 李梦莎, 许嘉阳, 张斌, 崔光周, 姜瑛. 烟草根际促生菌的筛选鉴定及促生性能研究[J]. 生物技术通报, 2025, 41(3): 271-281. |
| [5] | 葛仕杰, 刘怡德, 张华东, 宁强, 朱展望, 王书平, 刘易科. 小麦蛋白质二硫键异构酶基因家族的鉴定与表达[J]. 生物技术通报, 2025, 41(2): 85-96. |
| [6] | 张晓英, 毛咪, 王洪凤, 丁新华, 朱树伟, 石磊. 免疫诱抗剂ZNC对小麦赤霉病防治和产量的影响[J]. 生物技术通报, 2024, 40(8): 95-105. |
| [7] | 张娜, 刘梦楠, 屈展帆, 崔祎平, 倪嘉瑶, 王华忠. 小麦烯醇化酶基因ENO2的可变翻译分析和原核表达[J]. 生物技术通报, 2024, 40(5): 112-119. |
| [8] | 田姗姗, 黄诗宇, 杨天为, 高曼熔, 张尚文, 何龙飞, 张向军, 李婷, 石前. 高温干旱复合胁迫下铁皮石斛MYB基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(12): 145-159. |
| [9] | 牛德, 何跃辉. 季节性因素调控小麦开花时间的分子与表观遗传机制[J]. 生物技术通报, 2024, 40(10): 30-40. |
| [10] | 张怡, 张心如, 张金珂, 胡利宗, 上官欣欣, 郑晓红, 胡娟娟, 张聪聪, 穆桂清, 李成伟. 小麦镉胁迫响应基因TaMYB1的功能分析[J]. 生物技术通报, 2024, 40(1): 194-206. |
| [11] | 焦进兰, 王文文, 介欣芮, 王华忠, 岳洁瑜. 外源钙缓解小麦幼苗盐胁迫的作用机制[J]. 生物技术通报, 2024, 40(1): 207-221. |
| [12] | 常泸尹, 王中华, 李凤敏, 高梓源, 张辉红, 王祎, 李芳, 韩燕来, 姜瑛. 玉米根际多功能促生菌的筛选及其对冬小麦-夏玉米轮作体系产量提升效果[J]. 生物技术通报, 2024, 40(1): 231-242. |
| [13] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [14] | 韩志阳, 贾子苗, 梁秋菊, 王轲, 唐华丽, 叶兴国, 张双喜. 二套小麦-簇毛麦染色体附加系苗期耐盐性及籽粒硒和叶酸的含量[J]. 生物技术通报, 2023, 39(8): 185-193. |
| [15] | 任丽, 乔舒婷, 葛晨辉, 魏梓桐, 徐晨曦. 菠菜PSY基因家族的鉴定与表达分析[J]. 生物技术通报, 2023, 39(12): 169-178. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||