








生物技术通报 ›› 2024, Vol. 40 ›› Issue (1): 194-206.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0672
张怡1,2( ), 张心如1,2, 张金珂1,2, 胡利宗1,2, 上官欣欣1,2, 郑晓红1,2, 胡娟娟1,2, 张聪聪1,2, 穆桂清1,2, 李成伟3(
), 张心如1,2, 张金珂1,2, 胡利宗1,2, 上官欣欣1,2, 郑晓红1,2, 胡娟娟1,2, 张聪聪1,2, 穆桂清1,2, 李成伟3( )
)
收稿日期:2023-07-14
出版日期:2024-01-26
发布日期:2024-02-06
通讯作者:
李成伟,男,教授,研究方向:植物与微生物互作;E-mail: lichengweihist@163.com作者简介:张怡,女,博士,副教授,研究方向:植物与微生物互作;E-mail: yizhang0401@sina.com
基金资助:
ZHANG Yi1,2( ), ZHANG Xin-ru1,2, ZHANG Jin-ke1,2, HU Li-zong1,2, SHANGGUAN Xin-xin1,2, ZHENG Xiao-hong1,2, HU Juan-juan1,2, ZHANG Cong-cong1,2, MU Gui-qing1,2, LI Cheng-wei3(
), ZHANG Xin-ru1,2, ZHANG Jin-ke1,2, HU Li-zong1,2, SHANGGUAN Xin-xin1,2, ZHENG Xiao-hong1,2, HU Juan-juan1,2, ZHANG Cong-cong1,2, MU Gui-qing1,2, LI Cheng-wei3( )
)
Received:2023-07-14
Published:2024-01-26
Online:2024-02-06
摘要:
【目的】MYB家族成员在植物生长发育、生物胁迫和非生物胁迫等方面发挥重要调控作用,然而,小麦MYB转录因子在重金属镉(Cd)胁迫中的生物学功能研究较少。挖掘小麦Cd胁迫应答相关基因,阐明其在Cd胁迫中的生物学功能,为耐/低Cd小麦品种的选育奠定基础。【方法】构建Cd胁迫小麦根系酵母cDNA文库,筛选获得Cd胁迫应答基因,利用荧光定量PCR技术检测Cd胁迫条件下小麦不同组织中Cd胁迫应答基因的相对表达量;利用病毒诱导的基因沉默对该基因进行功能验证,检测对照和沉默植株的Cd含量、叶绿素含量、丙二醛(MDA)含量、超氧化物歧化酶(SOD)和过氧化物酶(POD)活力等生理生化指标。此外,对Cd胁迫应答基因及其在小麦和其他物种中的同源基因进行生物信息学分析(系统进化关系、序列特征和表达谱)。【结果】Cd胁迫主要抑制小麦根系发育,在添加0.002 mol/L Cd的酵母培养基上筛选Cd胁迫小麦根系酵母cDNA文库获得18个耐Cd转化子,其中5个转化子编码TaMYB1蛋白,转化TaMYB1酵母菌株在高Cd培养基中生长良好,而对照则明显受到抑制。RT-qPCR结果表明,TaMYB1在小麦幼苗中响应Cd胁迫。与对照相比,TaMYB1沉默植株中TaMYB1表达量明显下降,且根系和叶片Cd含量显著低于对照植株,叶绿素含量、SOD、POD活性高于对照植株,MDA含量则低于对照植株。同时,生物信息学分析发现,TaMYB1属于1R-MYB家族成员,具有高度保守的MYB和CC结构域,其包含7个外显子和6个内含子,在拔节期中期的花序和成熟期的花序表达最高,在灌浆前期种子和灌浆中后期种子中表达量最低。【结论】TaMYB1在小麦响应Cd胁迫应答中发挥重要作用。
张怡, 张心如, 张金珂, 胡利宗, 上官欣欣, 郑晓红, 胡娟娟, 张聪聪, 穆桂清, 李成伟. 小麦镉胁迫响应基因TaMYB1的功能分析[J]. 生物技术通报, 2024, 40(1): 194-206.
ZHANG Yi, ZHANG Xin-ru, ZHANG Jin-ke, HU Li-zong, SHANGGUAN Xin-xin, ZHENG Xiao-hong, HU Juan-juan, ZHANG Cong-cong, MU Gui-qing, LI Cheng-wei. Functional Analysis of TaMYB1 Gene in Wheat Under Cadmium Stress[J]. Biotechnology Bulletin, 2024, 40(1): 194-206.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| TaMYB1-F | GTGAGTAAGGTTACCGAACTCCAAATGCAAATTGAG |
| TaMYB1-R | CGTGAGCTCGGTACCGGAGACGAGGCTAGCTCCGGC |
| TaMYB1-qPCR-F | AATTGAGGTCCAGAGACGACTG |
| TaMYB1-qPCR-R | GGTACTTCCCCTGGGCTTCG |
| TaActin-qPCR-F | CCAGGTATCGCTGACCGTAT |
| TaActin-qPCR-R | GCTGAGTGAGGCTAGGATGG |
表1 引物序列
Table 1 Primer sequences
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| TaMYB1-F | GTGAGTAAGGTTACCGAACTCCAAATGCAAATTGAG |
| TaMYB1-R | CGTGAGCTCGGTACCGGAGACGAGGCTAGCTCCGGC |
| TaMYB1-qPCR-F | AATTGAGGTCCAGAGACGACTG |
| TaMYB1-qPCR-R | GGTACTTCCCCTGGGCTTCG |
| TaActin-qPCR-F | CCAGGTATCGCTGACCGTAT |
| TaActin-qPCR-R | GCTGAGTGAGGCTAGGATGG |

图1 Cd胁迫小麦表型检测 A:Cd胁迫小麦植株表型;B:Cd胁迫小麦植株根长;C:Cd胁迫小麦植株地上部长度;D:Cd胁迫小麦植株生物量。所有数据均重复3次以上,**代表在P=0.01水平上差异显著
Fig. 1 Phenotype analysis of wheat plant under Cd stress A: Phenotype of wheat plants under Cd stress. B: Root length of wheat plants under Cd stress. C: Shoot length of wheat plants under Cd stress. D: Total biomass of wheat plants under Cd stress. ** indicates significant difference from control plants at P=0.01
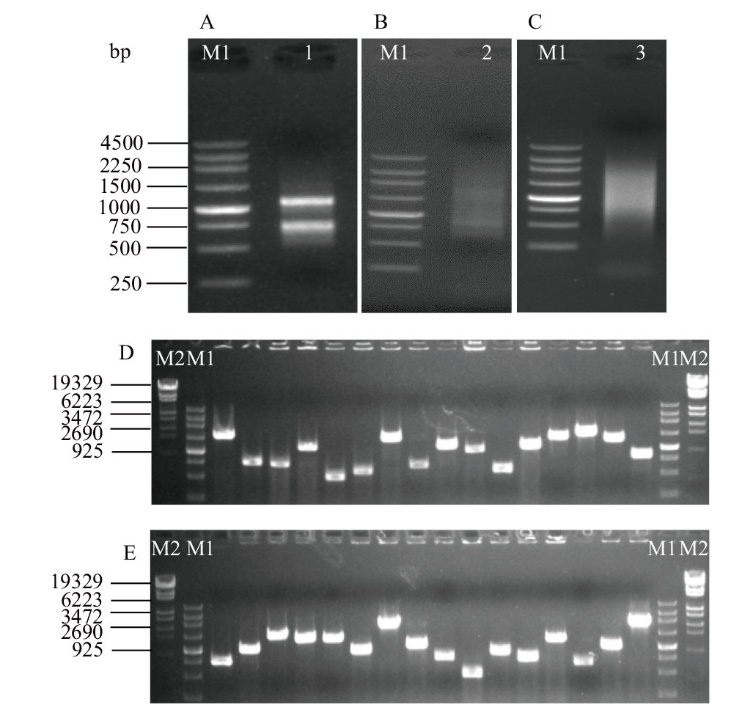
图2 Cd胁迫小麦根系的酵母双杂交文库质量检测 A:Cd胁迫小麦根系总RNA;B:利用SMART技术获得高质量cDNA; C:柱层析纯化后的cDNA;D, E:酵母文库插入片段PCR检测
Fig. 2 Quality detection of yeast two-hybrid library of wheat roots under Cd stress A: Total RNA of wheat roots under Cd stress. B: High-quality cDNA generated using SMART cDNA synthesis technology. C: Purified cDNA with column chromatography. D, E: PCR analysis of inserted clones from the yeast library

图3 转化子在添加不同Cd浓度的培养基上生长情况 A-C:在添加0.002、0.003和0.004 mol/L CdCl2的酵母培养基上筛选酵母文库的结果
Fig. 3 Transformants with the medium containing different cadmium concentration A-C: Refer to the screening results of yeast two-hybrid library in the yeast medium containing 0.002, 0.003 and 0.004 mol/L CdCl2
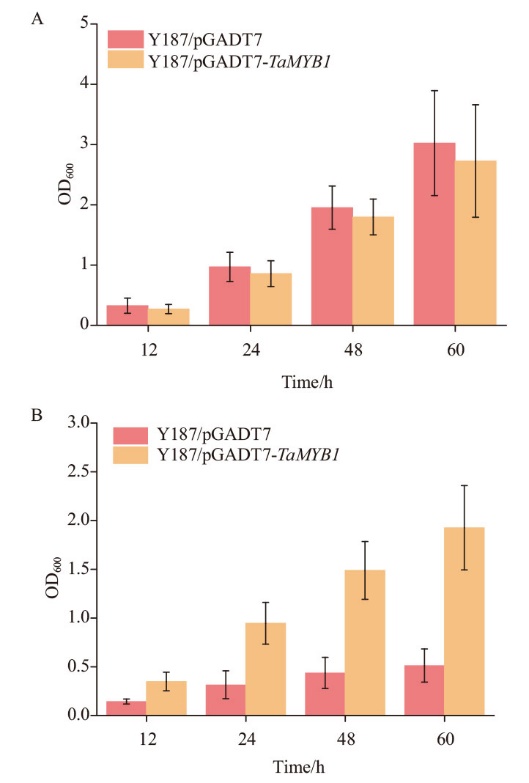
图4 酵母菌落在添加CdCl2的SD-Leu培养基中的生长曲线 A:酵母菌在正常条件下的生长曲线;B:酵母菌在高Cd培养基中的生长曲线
Fig. 4 Growth curves of yeast colonies in the SD-Leu medium added with CdCl2 A: Growth curves of yeast transformants growing in SD-Leu medium. B: Growth curves of yeast transformants growing in 0.004 mol/L CdCl2 medium
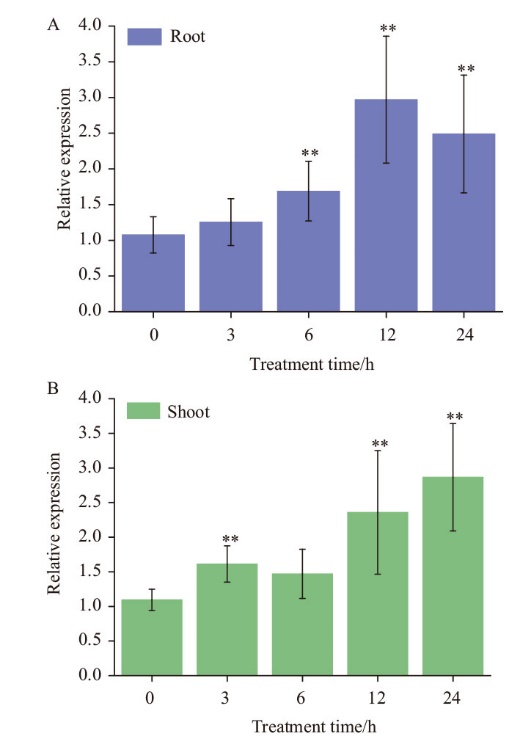
图5 TaMYB1响应Cd胁迫表达量分析 A:根系;B:地上部分。所有数据均重复3次以上,**代表在P=0.01水平上差异显著,下同
Fig. 5 Expression analysis of TaMYB1 under Cd stress A: Root. B: Shoot. ** indicates significant difference from control plants at P=0.01, the same below
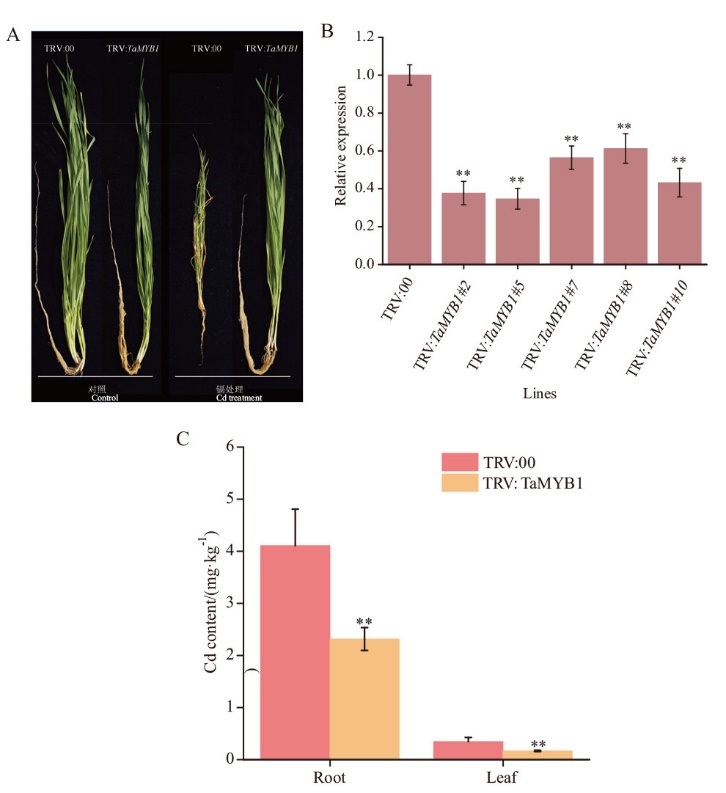
图6 沉默TaMYB1基因增强小麦对Cd耐受 A:TaMYB1基因沉默和对照植株在正常条件和Cd胁迫后的表型; B:TaMYB1沉默植株中TaMYB1基因表达量检测;C:TaMYB1基因沉默和对照植株Cd胁迫后植株的Cd含量
Fig. 6 Silencing of TaMYB1 enhanced wheat tolerance to cadmium A: Phenotypes of TaMYB1 gene-silenced and control plants under normal conditions and cadmium stress. B: Relative expression level of TaMYB1 gene in gene-silenced plants. C: Cadmium content of TaMYB1 gene-silenced and control plants after cadmium stress

图7 TaMYB1沉默和对照植株在正常条件和Cd胁迫后的生理指标检测 A-D中数据均取自代表小麦叶片
Fig. 7 Physiological index detection of TaMYB1 gene-silenced and control plants under normal conditions and cadmium stress The data in A-D were all taken from wheat leaves

图9 TaMYB1及其小麦同源基因的序列特征及系统进化关系 A:TaMYB1及其同源蛋白的系统进化分析;B:TaMYB1及其同源蛋白的保守基序;C:TaMYB1及其同源基因的结构;D:保守基序1的序列标识图;E:保守基序2的序列标识图
Fig. 9 Sequence features and phylogenetic relationships of TaMYB1 and its homologous genes in wheat A: Phylogenetic tree of TaMYB1 and its homologous proteins was analyzed. B: Conserved motifs of TaMYB1 and its homologous protein. C: Structures of TaMYB1 and its homologous genes. D: The sequence logo of the conserved motif 1. E: The sequence logo of the conserved motif 2
| [1] | 中华人民共和国环境保护部, 国土资源部. 全国土壤污染状况调查公报[R]. 北京: 中华人民共和国环境保护部, 国土资源部, 2014. |
| Ministry of Environmental Protection of PRC, Ministry of Land and Resources of PRC. Report on the national general survey of soil contamination[R]. Beijing: Ministry of Environmental Protection of PRC, Ministry of Land and Resources of PRC, 2014. | |
| [2] | 尚二萍, 许尔琪, 张红旗, 等. 中国粮食主产区耕地土壤重金属时空变化与污染源分析[J]. 环境科学, 2018, 39(10): 4670-4683. |
| Shang EP, Xu EQ, Zhang HQ, et al. Spatial-temporal trends and pollution source analysis for heavy metal contamination of cultivated soils in five major grain producing regions of China[J]. Environ Sci, 2018, 39(10): 4670-4683. | |
| [3] |
Raza A, Habib M, Kakavand SN, et al. Phytoremediation of cadmium: physiological, biochemical, and molecular mechanisms[J]. Biology, 2020, 9(7): 177.
doi: 10.3390/biology9070177 URL |
| [4] |
Rizwan M, Rubina Gilani S, Iqbal Durani A, et al. Materials diversity of hydrogel: synthesis, polymerization process and soil conditioning properties in agricultural field[J]. J Adv Res, 2021, 33: 15-40.
doi: 10.1016/j.jare.2021.03.007 pmid: 34603776 |
| [5] | 王怡雯, 芮玉奎, 李中阳, 等. 冬小麦吸收重金属特征及与影响因素的定量关系[J]. 环境科学, 2020, 41(3): 1482-1490. |
| Wang YW, Rui YK, Li ZY, et al. Characteristics of heavy metal absorption by winter wheat and its quantitative relationship with influencing factors[J]. Environ Sci, 2020, 41(3): 1482-1490. | |
| [6] | 赵多勇, 魏益民, 魏帅, 等. 区域小麦籽粒重金属分布及暴露评估[J]. 中国粮油学报, 2016, 31(7): 6-10, 18. |
| Zhao DY, Wei YM, Wei S, et al. Spatial distribution of heavy metal in wheat kernal and dietary exposure assessment of local residents in an industrial area[J]. J Chin Cereals Oils Assoc, 2016, 31(7): 6-10, 18. | |
| [7] | 肖冰, 薛培英, 韦亮, 等. 基于田块尺度的农田土壤和小麦籽粒镉砷铅污染特征及健康风险评价[J]. 环境科学, 2020, 41(6): 2869-2877. |
| Xiao B, Xue PY, Wei L, et al. Characteristics of Cd, As, and Pb in soil and wheat grains and health risk assessment of grain-Cd/As/Pb on the field scale[J]. Environ Sci, 2020, 41(6): 2869-2877. | |
| [8] |
Shi TR, Zhang YY, Gong YW, et al. Status of cadmium accumulation in agricultural soils across China(1975-2016): From temporal and spatial variations to risk assessment[J]. Chemosphere, 2019, 230: 136-143.
doi: 10.1016/j.chemosphere.2019.04.208 URL |
| [9] |
Hu YA, Cheng HF, Tao S. The challenges and solutions for cadmium-contaminated rice in China: A critical review[J]. Environ Int, 2016, 92/93: 515-532.
doi: 10.1016/j.envint.2016.04.042 URL |
| [10] | 王兴利, 王晨野, 吴晓晨, 等. 重金属污染土壤修复技术研究进展[J]. 化学与生物工程, 2019, 36(2): 1-7, 11. |
| Wang XL, Wang CY, Wu XC, et al. Research progress in remediation technology of heavy metal contaminated soil[J]. Chem Bioeng, 2019, 36(2): 1-7, 11. | |
| [11] |
Dinh N, van der Ent A, Mulligan DR, et al. Zinc and lead accumulation characteristics and in vivo distribution of Zn2+ in the hyperaccumulator Noccaea caerulescens elucidated with fluorescent probes and laser confocal microscopy[J]. Environ Exp Bot, 2018, 147: 1-12.
doi: 10.1016/j.envexpbot.2017.10.008 URL |
| [12] |
Wen X, Ding Y, Tan Z, et al. Identification and characterization of cadmium stress-related LncRNAs from Betula platyphylla[J]. Plant Sci, 2020, 299: 110601.
doi: 10.1016/j.plantsci.2020.110601 URL |
| [13] |
Chen L, Shi S, Jiang N, et al. Genome-wide analysis of long non-coding RNAs affecting roots development at an early stage in the rice response to cadmium stress[J]. BMC Genomics, 2018, 19(1): 460.
doi: 10.1186/s12864-018-4807-6 pmid: 29902991 |
| [14] |
Meng JG, Zhang XD, Tan SK, et al. Genome-wide identification of Cd-responsive NRAMP transporter genes and analyzing expression of NRAMP 1 mediated by miR167 in Brassica napus[J]. Biometals, 2017, 30(6): 917-931.
doi: 10.1007/s10534-017-0057-3 pmid: 28993932 |
| [15] |
Ding YF, Gong SH, Wang Y, et al. microRNA166 modulates cadmium tolerance and accumulation in rice[J]. Plant Physiol, 2018, 177(4): 1691-1703.
doi: 10.1104/pp.18.00485 pmid: 29925586 |
| [16] |
Wang NH, Zhou XY, Shi SH, et al. An miR156-regulated nucleobase-ascorbate transporter 2 confers cadmium tolerance via enhanced anti-oxidative capacity in barley[J]. J Adv Res, 2023, 44: 23-37.
doi: 10.1016/j.jare.2022.04.001 URL |
| [17] |
Han YY, Fan TT, Zhu XY, et al. WRKY12 represses GSH1 expression to negatively regulate cadmium tolerance in Arabidopsis[J]. Plant Mol Biol, 2019, 99(1): 149-159.
doi: 10.1007/s11103-018-0809-7 |
| [18] |
Li GZ, Zheng YX, Liu HT, et al. WRKY74 regulates cadmium tolerance through glutathione-dependent pathway in wheat[J]. Environ Sci Pollut Res Int, 2022, 29(45): 68191-68201.
doi: 10.1007/s11356-022-20672-6 |
| [19] |
Wang LJ, Lu WX, Ran LY, et al. R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa[J]. Plant J, 2019, 99(4): 733-751.
doi: 10.1111/tpj.v99.4 URL |
| [20] |
Pratyusha DS, Sarada DVL. MYB transcription factors-master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses[J]. Plant Cell Rep, 2022, 41(12): 2245-2260.
doi: 10.1007/s00299-022-02927-1 pmid: 36171500 |
| [21] |
Yang J, Chen Y, Xiao Z, et al. Multilevel regulation of anthocyanin-promoting R2R3-MYB transcription factors in plants[J]. Front Plant Sci, 2022, 13: 1008829.
doi: 10.3389/fpls.2022.1008829 URL |
| [22] |
Yin Y, Guo C, Shi HY, et al. Genome-wide comparative analysis of the R2R3-MYB gene family in five solanaceae species and identification of members regulating carotenoid biosynthesis in wolfberry[J]. Int J Mol Sci, 2022, 23(4): 2259.
doi: 10.3390/ijms23042259 URL |
| [23] |
Dubos C, Stracke R, Grotewold E et al. MYB transcription factors in Arabidopsis[J]. Trends Plant Sci, 2010, 15(10): 573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [24] |
Hu SB, Yu Y, Chen QH, et al. OsMYB45 plays an important role in rice resistance to cadmium stress[J]. Plant Sci, 2017, 264: 1-8.
doi: 10.1016/j.plantsci.2017.08.002 URL |
| [25] |
Zhang P, Wang RL, Ju Q, et al. The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation[J]. Plant Physiol, 2019, 180(1): 529-542.
doi: 10.1104/pp.18.01380 pmid: 30782964 |
| [26] |
Zhu S, Shi W, Jie Y, et al. A MYB transcription factor, BnMYB2, cloned from ramie(Boehmeria nivea)is involved in cadmium tolerance and accumulation[J]. PLoS One, 2020, 15(5): e0233375.
doi: 10.1371/journal.pone.0233375 URL |
| [27] |
Tiwari P, Indoliya Y, Chauhan AS et al. Over-expression of rice R1-type MYB transcription factor confers different abiotic stress tolerance in transgenic Arabidopsis[J]. Ecotoxicol Environ Saf, 2020, 206: 111361.
doi: 10.1016/j.ecoenv.2020.111361 URL |
| [28] | 宋红改, 蒋晶, 乔桂荣, 等. 利用酵母建立植物抗逆基因快速筛选体系[J]. 浙江林学院学报, 2010, 27(6): 890-895. |
| Song HG, Jiang J, Qiao GR, et al. Plant stress-resistant gene cloning for Na and Cd using an INVSc1 yeast[J]. J Zhejiang for Coll, 2010, 27(6): 890-895. | |
| [29] | 张怡, 刘坤, 曹鹏, 等. 一种快速高效提取酵母质粒的方法[J]. 食品与生物技术学报, 2013, 32(11): 1194-1198. |
| Zhang Y, Liu K, Cao P, et al. A fast, efficient and economical method for isolating plasmid from yeast[J]. J Food Sci Biotechnol, 2013, 32(11): 1194-1198. | |
| [30] | Zhang Y, Hu L, Yu D, et al. Integrative analysis of the wheat PHT1 gene family reveals a novel member involved in arbuscular mycorrhizal phosphate transport and immunity[J]. Cells, 2019, 8(5): E490. |
| [31] | 刘萍, 李明军. 植物生理实验技术[M]. 第2版. 北京: 科学出版社, 2008. |
| Liu P, Li MJ. Plant physiology experimental technology[M]. 2nd ed. Beijing: Science Press, 2008. | |
| [32] |
Mistry J, Chuguransky S, Williams L, et al. Pfam: The protein families database in 2021[J]. Nucleic Acids Res, 2021, 49(D1): D412-D419.
doi: 10.1093/nar/gkaa913 pmid: 33125078 |
| [33] |
Li KB. ClustalW-MPI: ClustalW analysis using distributed and parallel computing[J]. Bioinformatics, 2003, 19(12): 1585-1586.
doi: 10.1093/bioinformatics/btg192 URL |
| [34] |
Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [35] |
Letunic I, Bork JP. Interactive tree of life(iTOL)v5: an online tool for phylogenetic tree display and annotation[J]. Nucleic Acids Res, 2021, 49(W1): W293-W296.
doi: 10.1093/nar/gkab301 URL |
| [36] |
Hu B, Jin J, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296-1297.
doi: 10.1093/bioinformatics/btu817 pmid: 25504850 |
| [37] |
Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37: W202-W208.
doi: 10.1093/nar/gkp335 URL |
| [38] |
Crooks GE, Hon G, Chandonia JM, et al. WebLogo: a sequence logo generator[J]. Genome Res, 2004, 14(6): 1188-1190.
doi: 10.1101/gr.849004 pmid: 15173120 |
| [39] |
Pingault L, Choulet F, Alberti A, et al. Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome[J]. Genome Biol, 2015, 16(1): 29.
doi: 10.1186/s13059-015-0601-9 URL |
| [40] |
Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks[J]. Nat Protoc, 2012, 7(3): 562-578.
doi: 10.1038/nprot.2012.016 pmid: 22383036 |
| [41] |
Ning WS, Wei YX, Gao LT, et al. HemI 2.0: an online service for heatmap illustration[J]. Nucleic Acids Res, 2022, 50(W1): W405-W411.
doi: 10.1093/nar/gkac480 pmid: 35670661 |
| [42] |
Zadoks JC, Chang TT, Konzak CF. A decimal code for the growth stages of cereals[J]. Weed Res, 1974, 14(6): 415-421.
doi: 10.1111/wre.1974.14.issue-6 URL |
| [43] |
Shan TL, Rong W, Xu HJ, et al. The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes[J]. Sci Rep, 2016, 6(1): 1-14.
doi: 10.1038/s41598-016-0001-8 |
| [44] |
Hawku MD, He FX, Bai XX, et al. A R2R3 MYB transcription factor, TaMYB391, is positively involved in wheat resistance to Puccinia striiformis f. sp. tritici[J]. Int J Mol Sci, 2022, 23(22): 14070.
doi: 10.3390/ijms232214070 URL |
| [45] |
Wei XN, Shan TL, Hong YT, et al. TaPIMP2, a pathogen-induced MYB protein in wheat, contributes to host resistance to common root rot caused by Bipolaris sorokiniana[J]. Sci Rep, 2017, 7(1): 1754.
doi: 10.1038/s41598-017-01918-7 |
| [46] | 何冠谛. 低积累Cd型马铃薯相关基因挖掘及耐镉分子机制研究[D]. 贵阳: 贵州大学, 2021. |
| He GD. Study on gene mining and molecular mechanism of cadmium tolerance in low accumulation cd potato[D]. Guiyang: Guizhou University, 2021. | |
| [47] | 汪怡文. 转录组和代谢组整合分析冬小麦镉胁迫响应的关键代谢通路[D]. 武汉: 华中农业大学, 2021. |
| Wang YW. transcriptome and metabolonmics integrated analysis of key metabolic pathways in response to cadmium stress in winter wheat[D]. Wuhan: Huazhong Agricultural University, 2021. | |
| [48] |
Niu M, Bao C, Zhan J, et al. Plasma membrane-localized protein BcHIPP16 promotes the uptake of copper and cadmium in planta[J]. Ecotoxicol Environ Saf, 2021, 227: 112920.
doi: 10.1016/j.ecoenv.2021.112920 URL |
| [49] |
Martel AB, Taylor AE, Qaderi MM. Individual and interactive effects of temperature and light intensity on canola growth, physiological characteristics and methane emissions[J]. Plant Physiol Biochem, 2020, 157: 160-168.
doi: 10.1016/j.plaphy.2020.10.016 URL |
| [50] | 葛依立, 陈心胜, 黄道友, 等. 湿地植物水蓼(Polygonum hydropiper L.)对镉的富集特征及生理响应[J]. 生态毒理学报, 2020, 15(2): 195-200. |
| Ge YL, Chen XS, Huang DY, et al. Accumulation characteristics and physiological responses of the wetland plant, Polygonum hydropiper L. to cadmium[J]. Asian J Ecotoxicol, 2020, 15(2): 190-200. | |
| [51] | 史怀宇. LcAPX基因对植物镉耐受的影响及水稻组织培养条件的优化[D]. 天津: 天津大学, 2018. |
| Shi HY. Effects of LcAPX on plant cadmium resistance and optimizing of rice callus culture condition[D]. Tianjin: Tianjin University, 2018. | |
| [52] | 黄云. 马铃薯WRKY6基因增强拟南芥镉耐受性功能验证[D]. 贵阳: 贵州大学, 2022. |
| Huang Y. Function validation of Arabidopsis thaliana cadmium tolerance enhanced by potato WRKY6[D]. Guiyang: Guizhou University, 2022. |
| [1] | 焦进兰, 王文文, 介欣芮, 王华忠, 岳洁瑜. 外源钙缓解小麦幼苗盐胁迫的作用机制[J]. 生物技术通报, 2024, 40(1): 207-221. |
| [2] | 常泸尹, 王中华, 李凤敏, 高梓源, 张辉红, 王祎, 李芳, 韩燕来, 姜瑛. 玉米根际多功能促生菌的筛选及其对冬小麦-夏玉米轮作体系产量提升效果[J]. 生物技术通报, 2024, 40(1): 231-242. |
| [3] | 何思成, 张紫瑗, 韩雨晴, 苗琳, 张翠英, 于爱群. 解脂耶氏酵母细胞工厂生产多不饱和脂肪酸的研究进展[J]. 生物技术通报, 2024, 40(1): 72-85. |
| [4] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [5] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [6] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [7] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [8] | 徐发迪, 徐康, 孙东明, 李萌蕾, 赵建志, 鲍晓明. 基于杨木(Populus sp.)的二代燃料乙醇技术研究进展[J]. 生物技术通报, 2023, 39(9): 27-39. |
| [9] | 赵思佳, 王晓璐, 孙纪录, 田健, 张杰. 代谢工程改造毕赤酵母生产赤藓糖醇[J]. 生物技术通报, 2023, 39(8): 137-147. |
| [10] | 韩志阳, 贾子苗, 梁秋菊, 王轲, 唐华丽, 叶兴国, 张双喜. 二套小麦-簇毛麦染色体附加系苗期耐盐性及籽粒硒和叶酸的含量[J]. 生物技术通报, 2023, 39(8): 185-193. |
| [11] | 徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219. |
| [12] | 宋志忠, 徐维华, 肖慧琳, 唐美玲, 陈景辉, 管雪强, 刘万好. 酿酒葡萄铁调节转运蛋白基因VvIRT1的克隆、表达与功能[J]. 生物技术通报, 2023, 39(8): 234-240. |
| [13] | 李文辰, 刘鑫, 康越, 李伟, 齐泽铮, 于璐, 王芳. TRV病毒诱导大豆基因沉默体系优化及应用[J]. 生物技术通报, 2023, 39(7): 143-150. |
| [14] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [15] | 李雨真, 梅天秀, 李治文, 王淇, 李俊, 邹岳, 赵心清. 红酵母基因组和代谢工程改造研究进展[J]. 生物技术通报, 2023, 39(7): 67-79. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||