








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 66-73.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1133
豆硕1( ), 丁若羲1, 孙星2, 郭文静1, 孔文慧1, 袁静贤1, 张冬梅1, 王省芬1, 马峙英1, 吴金华1(
), 丁若羲1, 孙星2, 郭文静1, 孔文慧1, 袁静贤1, 张冬梅1, 王省芬1, 马峙英1, 吴金华1( ), 吴立柱1(
), 吴立柱1( )
)
收稿日期:2024-11-24
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
吴立柱,男,博士,副教授,研究方向 :棉花遗传育种和作物雄性不育;E-mail: wulizhu2008@163.com;作者简介:豆硕,男,硕士研究生,研究方向 :棉花遗传育种和作物雄性不育;E-mail: 1366761040@qq.com
基金资助:
DOU Shuo1( ), DING Ruo-xi1, SUN Xing2, GUO Wen-jing1, KONG Wen-hui1, YUAN Jing-xian1, ZHANG Dong-mei1, WANG Xing-fen1, MA Zhi-ying1, WU jin-hua1(
), DING Ruo-xi1, SUN Xing2, GUO Wen-jing1, KONG Wen-hui1, YUAN Jing-xian1, ZHANG Dong-mei1, WANG Xing-fen1, MA Zhi-ying1, WU jin-hua1( ), WU Li-zhu1(
), WU Li-zhu1( )
)
Received:2024-11-24
Published:2025-12-26
Online:2026-01-06
摘要:
目的 构建并验证一种可视化筛选转基因通用载体pCamRUBY,以期提高植物遗传转化效率。 方法 在以RUBY作为报告基因的基础上利用2A技术构建成一个通用表达载体pCamRUBY,以Spe和Kan抗性基因分别构建验证载体pCamSpeRUBY和pCamKanRUBY,并分别以拟南芥和棉花为材料进行遗传转化验证和色素含量鉴定。 结果 该载体具有可视化程度高,载体序列小,内含由Sal I、Apa I、Xba I、Sac I和Spe I组成的MCS位点和T2A(optimized)序列等特点。分别获得可裸眼观察筛选的具有红色表型性状的转基因拟南芥和棉花阳性植株或组织,并可得到稳定遗传后代。 结论 该通用表达载体pCamRUBY可直接连接外源基因正常表达、可稳定遗传且可直接裸眼观察筛选的红色性状转基因阳性植株,且不影响包含叶绿素含量在内的其他表型性状。
豆硕, 丁若羲, 孙星, 郭文静, 孔文慧, 袁静贤, 张冬梅, 王省芬, 马峙英, 吴金华, 吴立柱. 一种可视化筛选转基因通用载体pCamRUBY的构建和应用研究[J]. 生物技术通报, 2025, 41(12): 66-73.
DOU Shuo, DING Ruo-xi, SUN Xing, GUO Wen-jing, KONG Wen-hui, YUAN Jing-xian, ZHANG Dong-mei, WANG Xing-fen, MA Zhi-ying, WU jin-hua, WU Li-zhu. Construction and Application Study of a General Vector pCamRUBY for Visual Screening of Transgenes[J]. Biotechnology Bulletin, 2025, 41(12): 66-73.
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| RUBYF | CCGTGCACATCAACGACACC |
| RUBYR | GGAGGTGAACTTGTAGGAGC |
| AtActinF | CTGGTGATGGTGTGTCTCACAC |
| AtActinR | GCGATCCAGACACTGTACTTCC |
| GhActinF | GATCCTTCCTGATATCCACATCG |
| GhActinR | GGCACACTGGTGTTATGGTTGGG |
表1 引物名称及序列信息
Table 1 Primers' names and sequence information
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| RUBYF | CCGTGCACATCAACGACACC |
| RUBYR | GGAGGTGAACTTGTAGGAGC |
| AtActinF | CTGGTGATGGTGTGTCTCACAC |
| AtActinR | GCGATCCAGACACTGTACTTCC |
| GhActinF | GATCCTTCCTGATATCCACATCG |
| GhActinR | GGCACACTGGTGTTATGGTTGGG |
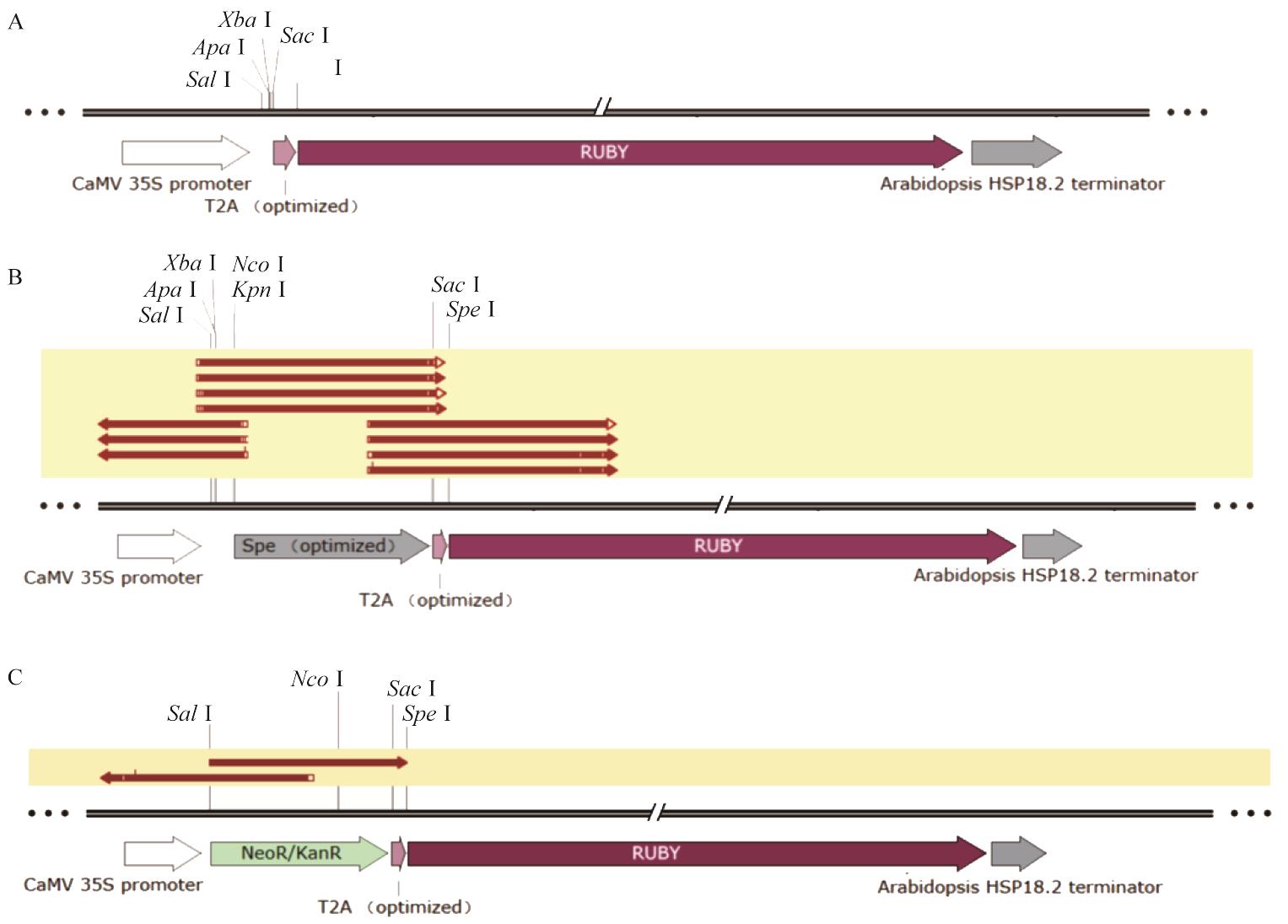
图1 载体结果示意图和测序比对结果A:pCamRUBY载体图谱;B:pCamSpeRUBY载体图谱和测序比对结果;C:pCamKanRUBY载体图谱和测序比对结果
Fig. 1 Schematic diagram of vector results and sequencing alignment resultsA: pCamRUBY vector profile. B: pCamSpeRUBY vector profile and sequencing alignment results. C: pCamKanRUBY vector profile and sequencing alignment results

图2 转基因的拟南芥的筛选A:利用Spe抗性筛选获得的红色拟南芥植株;B:营养土直接播种目测筛选的红色性状拟南芥植株;C:T2代pCamSpeRUBY转基因拟南芥植株;箭头所示为转基因阳性植株;标尺为1 cm
Fig. 2 Transgenic Arabidopsisthaliana screeningA: Red Arabidopsis were obtained through Spe resistance screening. B: Red Arabidopsis were directly screened with naked eye on nutrient soil. C: T2 generation pCamSpeRUBY transgenic Arabidopsis. The arrow indicates the transgenic positive plants. Bar=1 cm

图3 转基因拟南芥表型观察A:野生型和转基因拟南芥花序表型观察;B:野生型和转基因拟南芥抽薹期表型观察;C:野生型和转基因拟南芥(T3代)成苗单株表型观察;D:野生型和转基因拟南芥种子表型观察;E:转基因拟南芥RUBY基因的PCR电泳结果;F:转基因拟南芥AtActin基因的PCR电泳结果。M:DNA maker DL2000;1-2:抗性筛选获得的转基因拟南芥植株;3-5:直接播种获得的转基因植株;P:质粒pCamRUBY;WT:野生型对照拟南芥植株;c:空白对照;标尺为1 cm
Fig. 3 Phenotype of transgenic A. thalianaA: Phenotype of wild type and transgenic Arabidopsis inflorescence. B: Phenotype of bolting wild type and transgenic Arabidopsis. C: Phenotype of wild type and transgenic Arabidopsis plants. D: Phenotype of wild type and transgenic Arabidopsis seeds. E: PCR identification of RUBY gene in transgenic Arabidopsis. F: PCR identification of AtActin gene in in transgenic Arabidopsis. M: DNA maker DL2000. 1-2: Transgenic Arabidopsis plants obtained through resistance screening. 3-5: Transgenic Arabidopsis plants obtained by direct seeding. P: Plasmid pCamRUBY. WT: Wild-type control Arabidopsis plant. c: Blank control. Bar=1 cm
荚果编号 No. of pod | 种子数 Number of seeds | 红色种子数 Number of red seeds | 黄褐色种子数 Number of tawny seeds |
|---|---|---|---|
| 1 | 28 | 21 | 7 |
| 2 | 22 | 17 | 5 |
| 3 | 23 | 18 | 5 |
| 4 | 32 | 25 | 7 |
| 5 | 20 | 15 | 5 |
| 6 | 19 | 15 | 4 |
| 7 | 25 | 18 | 7 |
| 8 | 29 | 21 | 8 |
| 9 | 24 | 18 | 6 |
| 10 | 26 | 19 | 7 |
| Total | 248 | 187 | 61 |
表2 转基因拟南芥种子分离比统计
Table 2 Separation ratio statistics of transgenic Arabidopsis seeds
荚果编号 No. of pod | 种子数 Number of seeds | 红色种子数 Number of red seeds | 黄褐色种子数 Number of tawny seeds |
|---|---|---|---|
| 1 | 28 | 21 | 7 |
| 2 | 22 | 17 | 5 |
| 3 | 23 | 18 | 5 |
| 4 | 32 | 25 | 7 |
| 5 | 20 | 15 | 5 |
| 6 | 19 | 15 | 4 |
| 7 | 25 | 18 | 7 |
| 8 | 29 | 21 | 8 |
| 9 | 24 | 18 | 6 |
| 10 | 26 | 19 | 7 |
| Total | 248 | 187 | 61 |
图4 棉花遗传转化愈伤和植株表型观察和阳性植株PCR鉴定A:转基因棉花的愈伤组织呈现红色性状;B:转基因棉花阳性植株幼苗呈现红色性状;C:转基因棉花阳性植株成苗呈现红色性状;D:野生型和T1代转基因棉花表型观察;E:转基因棉花RUBY基因的PCR电泳结果;F:转基因棉花GhActin基因的PCR电泳结果。M:DNA maker DL2000;1-3:T0代转基因棉花植株;P:质粒pCamRUBY;WT:野生型对照棉花植株;c:空白对照;标尺为1 cm
Fig. 4 Phenotypic observation of genetically transformed cotton calli and plants and PCR identifications of positive plantsA: Red transgenic cotton callus. B: Red transgenic cotton seedlings. C: Red transgenic cotton plants. D: Phenotype of wild type and T1 transgenic plants. E: PCR identification of RUBY gene in transgenic cotton. F: PCR identification of GhActin gene in transgenic cotton. M: DNA marker DL2000; 1-3: T0 transgenic cotton plants; 4-6: T1 transgenic cotton plants; P: plasmid pCamRUBY; WT: wild-type cotton J668; c: blank control; Bar=1 cm
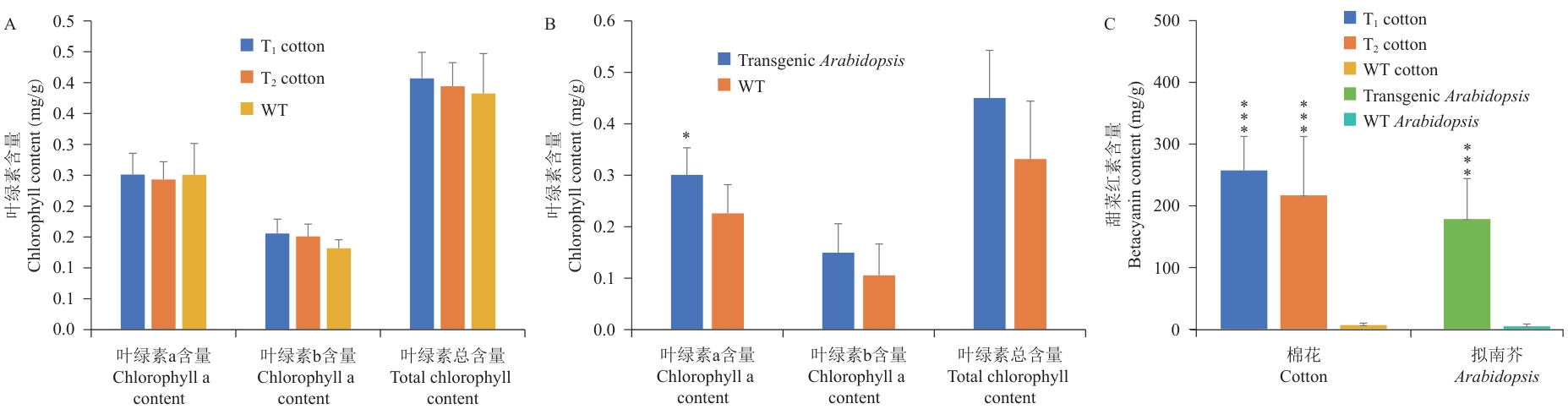
图5 转基因棉花和拟南芥的叶绿素和甜菜红素的含量检测A:转基因棉花叶绿素含量测定;B:转基因拟南芥叶绿素含量测定;C:转基因植株甜菜红素含量测定
Fig. 5 Content detection of chlorophyll and betacyanin in transgenic cotton and ArabidopsisA: Determination of chlorophyll content in transgenic cotton. B: Determination of chlorophyll content in transgenic A. thaliana. C: Determination of betacyanin content in transgenic plants. *P<0.05; ***P<0.001
| [1] | 翟琼, 陈容钦, 梁晓华, 等. 一种花生快速遗传转化方法的建立与应用 [J]. 植物学报, 2022, 57(3): 327-339. |
| Zhai Q, Chen RQ, Liang XH, et al. Establishment and application of a rapid genetic transformation method for peanut [J]. Chin Bull Bot, 2022, 57(3): 327-339. | |
| [2] | Tang YP, Shen XY, Deng X, et al. Establishment of an efficient Agrobacterium-mediated transformation system for chilli pepper and its application in genome editing [J]. Plant Biotechnol J, 2025, 23: 4752-4754. |
| [3] | 柴燕文, 马晖玲, 谢小冬, 等. pBI121-Lyz-GFP表达载体的构建及其在和田苜蓿愈伤组织中的转化 [J]. 农业生物技术学报, 2008, 16(5): 842-846. |
| Chai YW, Ma HL, Xie XD, et al. Construction of pBI121-lyz-GFP expression vector and its transformation in callus of Medicago sativa [J]. J Agric Biotechnol, 2008, 16(5): 842-846. | |
| [4] | 张献龙, 李涛, 孙济中. 抗生素对棉花愈伤组织诱导和生长的影响 [J]. 华中农业大学学报, 1996, 15(2): 123-126. |
| Zhang XL, Li T, Sun JZ. The effect of antibiotics on the callus induction and growth in upland cotton (Gossypium hirsutum L.) [J]. J Huazhong Cent China Agric Univ, 1996, 15(2): 123-126. | |
| [5] | 刘巧泉, 陈秀花, 王兴稳, 等. 一种快速检测转基因水稻中潮霉素抗性的简易方法 [J]. 农业生物技术学报, 2001, 9(3): 264-268, 308. |
| Liu QQ, Chen XH, Wang XW, et al. A rapid simple method of assaying hygromycin resistance in transgenic rice plants [J]. J Agric Biotechnol, 2001, 9(3): 264-268, 308. | |
| [6] | Borovinskaya MA, Shoji S, Fredrick K, et al. Structural basis for hygromycin B inhibition of protein biosynthesis [J]. RNA, 2008, 14(8): 1590-1599. |
| [7] | 肖国樱, 唐俐, 袁定阳, 等. 转Bar基因抗除草剂两系杂交早稻恢复系Bar 68-1的培育研究 [J]. 杂交水稻, 2007, 22(6): 57-61. |
| Xiao GY, Tang L, Yuan DY, et al. Studies on development of bar 68-1, a bar-transgenic restorer line with herbicide resistance of two-line early hybrid rice [J]. Hybrid Rice, 2007, 22(6): 57-61. | |
| [8] | Subach FV, Patterson GH, Manley S, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy [J]. Nat Methods, 2009, 6(2): 153-159. |
| [9] | 陶曼芝, 吴席, 朱香豫, 等. 拟南芥ProWRKY12-GUS转基因植株的构建及其染色分析 [J]. 合肥工业大学学报: 自然科学版, 2022, 45(9): 1248-1251. |
| Tao MZ, Wu X, Zhu XY, et al. Construction and staining analysis of Arabidopsis thaliana ProWRKY12-GUS transgenic plants [J]. J Hefei Univ Technol Nat Sci, 2022, 45(9): 1248-1251. | |
| [10] | 崔志芳, 邹玉红, 季爱云. 绿色荧光蛋白研究的三个里程碑——2008年诺贝尔化学奖简介 [J]. 自然杂志, 2008, 30(6): 324-328. |
| Cui ZF, Zou YH, Ji AY. Three landmarks during the research of the green fluorescent protein(GFP): a brief introduction to the 2008 Nobel prize in chemistry [J]. Chin J Nat, 2008, 30(6): 324-328. | |
| [11] | Magness ST, Bataller R, Yang L, et al. A dual reporter gene transgenic mouse demonstrates heterogeneity in hepatic fibrogenic cell populations [J]. Hepatology, 2004, 40(5): 1151-1159. |
| [12] | Uenaka H, Kadota A. Functional analyses of the Physcomitrella patens phytochromes in regulating chloroplast avoidance movement [J]. Plant J, 2007, 51(6): 1050-1061. |
| [13] | 朱向玲, 吴旭铭, 王卉卉, 等. 基于Cre/Loxp系统的Csf1r-CreERT2R26REYFP报告基因小鼠的构建及效率检测 [J]. 安徽医科大学学报, 2024, 59(7): 1175-1180. |
| Zhu XL, Wu XM, Wang HH, et al. Construction and efficiency detection of Csf1r-CreERT2 R26REYFP reporter gene mouse based on Cre/Loxp system [J]. Acta Univ Med Anhui, 2024, 59(7): 1175-1180. | |
| [14] | He YB, Zhang T, Sun H, et al. A reporter for noninvasively monitoring gene expression and plant transformation [J]. Hortic Res, 2020, 7(1): 152. |
| [15] | 黄晨, 张威, 任红旭. 植物中甜菜色素的研究进展 [J]. 西北植物学报, 2023, 43(12): 2149-2160. |
| Huang C, Zhang W, Ren HX. Research progress of betalain in plants [J]. Acta Bot Boreali Occidentalia Sin, 2023, 43(12): 2149-2160. | |
| [16] | Liu J, Li H, Hong CH, et al. Quantitative RUBY reporter assay for gene regulation analysis [J]. Plant Cell Environ, 2024, 47(10): 3701-3711. |
| [17] | Šola K, Gilchrist EJ, Ropartz D, et al. RUBY, a putative galactose oxidase, influences pectin properties and promotes cell-to-cell adhesion in the seed coat epidermis of Arabidopsis [J]. Plant Cell, 2019, 31(4): 809-831. |
| [18] | Wang D, Zhong Y, Feng B, et al. The RUBY reporter enables efficient haploid identification in maize and tomato [J]. Plant Biotechnol J, 2023, 21(8): 1707-1715. |
| [19] | 林凯, 张育华, 柴霞, 等. 发根农杆菌介导的星油藤毛状根遗传转化体系建立 [J]. 植物生理学报, 2023, 59(2): 373-382. |
| Lin K, Zhang YH, Chai X, et al. Establishment of an Agrobacterium rhizogenes-mediated genetic transformation system of hairy roots in Sacha inchi (Plukenetia volubilis L.) [J]. Plant Physiol J, 2023, 59(2): 373-382. | |
| [20] | 吴立柱, 安叶芝, 张洁, 等. 基于核糖体跳跃的多顺反子共表达技术在拟南芥上的应用 [J]. 农业生物技术学报, 2019, 27(7): 1141-1148. |
| Wu LZ, An YZ, Zhang J, et al. Application of ribosomal skipping based polycistronic co-expression technique in Arabidopsis thaliana [J]. J Agric Biotechnol, 2019, 27(7): 1141-1148. | |
| [21] | Momose F, Morikawa Y. Polycistronic expression of the influenza a virus RNA-dependent RNA polymerase by using the Thosea asigna virus 2A-like self-processing sequence [J]. Front Microbiol, 2016, 7: 288. |
| [22] | 吴立柱, 王省芬, 李喜焕, 等. 通用型植物表达载体pCamE的构建及功能验证 [J]. 农业生物技术学报, 2014, 22(6): 661-671. |
| Wu LZ, Wang XF, Li XH, et al. Construction and function identification of universal plant expression vector pCamE [J]. J Agric Biotechnol, 2014, 22(6): 661-671. | |
| [23] | 马峙英, 吴立柱, 王省芬, 等. 植物表达载体pCamE及其在基因转化中的应用: CN102816790A [P]. 2012-12-12. |
| Ma ZY, Wu LZ, Wang XF, et al. Plant expression vector pCamE and application thereof to gene transformation: CN102816790A [P]. 2012-12-12. | |
| [24] | Ali I, Sher H, Ali A, et al. Simplified floral dip transformation method of Arabidopsis thaliana [J]. J Microbiol Meth, 2022, 197: 106492. |
| [25] | 鲍红帅, 尚红燕, 王国宁, 等. 不同陆地棉品种愈伤组织诱导比较及遗传转化验证 [J]. 河北农业大学学报, 2023, 46(1): 31-36. |
| Bao HS, Shang HY, Wang GN, et al. Comparison of callus induction ability of different upland cotton varieties and verification of genetic transformation [J]. J Hebei Agric Univ, 2023, 46(1): 31-36. | |
| [26] | 段秋霞, 李定金, 段振华, 等. 红心火龙果酒颜色稳定性影响因素的研究 [J]. 食品研究与开发, 2020, 41(24): 130-136. |
| Duan QX, Li DJ, Duan ZH, et al. Study on the factors affecting the color stability of red heart pitaya wine [J]. Food Res Dev, 2020, 41(24): 130-136. | |
| [27] | Liu HQ, Zhao JY, Chen FF, et al. Improving Agrobacterium tumefaciens-mediated genetic transformation for gene function studies and mutagenesis in cucumber (Cucumis sativus L.) [J]. Genes, 2023, 14(3): 601. |
| [28] | 葛杰, 何玉池, 展帆, 等. 多倍体水稻DHF-CBS基因的抗旱性鉴定 [J]. 湖北农业科学, 2024, 63(9): 216-220. |
| Ge J, He YC, Zhan F, et al. Identification of drought resistance of polyploid Oryza sativa L. DHF-CBS gene [J]. Hubei Agric Sci, 2024, 63(9): 216-220. | |
| [29] | Bai MY, Yuan JH, Kuang HQ, et al. Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean [J]. Plant Biotechnol J, 2020, 18(3): 721-731. |
| [30] | Berthomieu P, Béclin C, Charlot F, et al. Routine transformation of rapid cycling cabbage (Brassica oleracea)—molecular evidence for regeneration of chimeras [J]. Plant Sci, 1994, 96(1/2): 223-235. |
| [31] | Mathews H, Dewey V, Wagoner W, et al. Molecular and cellular evidence of chimaeric tissues in primary transgenics and elimination of chimaerism through improved selection protocols [J]. Transgenic Res, 1998, 7(2): 123-129. |
| [32] | Faize M, Faize L, Burgos L. Using quantitative real-time PCR to detect chimeras in transgenic tobacco and apricot and to monitor their dissociation [J]. BMC Biotechnol, 2010, 10: 53. |
| [33] | Wu YH, Li J, Wang YL, et al. Development and application of a general plasmid reference material for GMO screening [J]. Plasmid, 2016, 87-88: 28-36. |
| [1] | 胡璐, 王凯, 徐婧仪, 叶丽慧, 王永飞, 王丽华, 李杰勤. 玉米和高粱遗传转化技术研究进展[J]. 生物技术通报, 2025, 41(9): 32-43. |
| [2] | 邓美壁, 严浪, 詹志田, 朱敏, 和玉兵. RUBY辅助的水稻高效CRISPR基因编辑[J]. 生物技术通报, 2025, 41(8): 65-73. |
| [3] | 裴景琪, 赵梦然, 黄晨阳, 邬向丽. 一个影响糙皮侧耳生长发育的功能基因的发现与验证[J]. 生物技术通报, 2025, 41(6): 327-334. |
| [4] | 杨朝结, 张兰, 陈红, 黄娟, 石桃雄, 朱丽伟, 陈庆富, 李洪有, 邓娇. 苦荞转录因子基因FtbHLH3调控类黄酮生物合成的功能鉴定[J]. 生物技术通报, 2025, 41(4): 134-144. |
| [5] | 文博霖, 万敏, 胡建军, 王克秀, 景晟林, 王心悦, 朱博, 唐铭霞, 李兵, 何卫, 曾子贤. 马铃薯川芋50遗传转化及基因编辑体系的建立[J]. 生物技术通报, 2025, 41(4): 88-97. |
| [6] | 张欢欢, 穆晓娅, 周静宜, 吕高培, 肖楠, 李敏, 郝曜山, 吴慎杰. 糜子高频胚胎发生品种遗传转化体系的建立[J]. 生物技术通报, 2025, 41(10): 164-174. |
| [7] | 王碧成, 景海青, 万坤, 张莹莹, 丁家豪, 李润植, 薛金爱, 张海平. 大豆BCAT基因家族鉴定及GmBCAT3在干旱胁迫中的功能分析[J]. 生物技术通报, 2025, 41(10): 196-209. |
| [8] | 李粘前, 李琛, 李淑婷, 马菊花, 景海青, 孙岩, 周雅莉, 薛金爱, 李润植. 油莎豆苹果酸酶(ME)全基因组鉴定及CeNAD-ME2功能分析[J]. 生物技术通报, 2025, 41(10): 210-221. |
| [9] | 宋倩娜, 段永红, 冯瑞云. CRISPR/Cas9介导的高效四倍体马铃薯试管薯基因编辑体系的建立[J]. 生物技术通报, 2024, 40(9): 33-41. |
| [10] | 张美玉, 赵玉斌, 王灵云, 宋元达, 赵新河, 任晓洁. 微藻破囊壶菌产功能性脂肪酸DHA研究进展[J]. 生物技术通报, 2024, 40(6): 81-94. |
| [11] | 梅显军, 宋慧洋, 李京昊, 梅超, 宋倩娜, 冯瑞云, 陈喜明. 马铃薯StDof5的克隆及表达分析[J]. 生物技术通报, 2024, 40(3): 181-192. |
| [12] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [13] | 周家燕, 邹建, 陈卫英, 吴一超, 陈奚潼, 王倩, 曾文静, 胡楠, 杨军. 植物多基因干扰载体体系构建与效用分析[J]. 生物技术通报, 2023, 39(1): 115-126. |
| [14] | 孙威, 张艳, 王聿晗, 徐僡, 徐小蓉, 鞠志刚. 马缨杜鹃Rd3GT1的克隆及对矮牵牛花色形成的影响[J]. 生物技术通报, 2022, 38(9): 198-206. |
| [15] | 陆新华, 孙德权, 张秀梅. 介孔硅纳米粒作为植物细胞转基因载体的研究[J]. 生物技术通报, 2022, 38(7): 194-204. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||