








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 201-213.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0507
收稿日期:2025-05-17
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
张文娥,女,博士,教授,研究方向 :观赏植物栽培生理;E-mail: agr.wezhang@gzu.edu.cn作者简介:杨晨欣,女,硕士研究生,研究方向 :美人蕉花色;E-mail: 1073491690@qq.com
基金资助:
YANG Chen-xin( ), LI Meng-xiu, JIANG Tang, ZHANG Wen-e(
), LI Meng-xiu, JIANG Tang, ZHANG Wen-e( ), PAN Xue-jun
), PAN Xue-jun
Received:2025-05-17
Published:2025-12-26
Online:2026-01-06
摘要:
目的 鉴定美人蕉(Canna indica)R2R3-MYB基因家族,分析其在不同花色中的表达模式,为美人蕉花色分子调控机制及新品种培育提供理论参考。 方法 以模式植物拟南芥R2R3-MYB蛋白序列为参考,结合HMM模型和BLAST检索从美人蕉基因组中鉴定R2R3-MYB基因家族成员,分析其理化性质、亚细胞定位、系统发育树、染色体分布、保守基序及启动子顺式作用元件;基于课题组前期8个美人蕉品种转录组数据,通过TBtools可视化分析获得其在盛花期花瓣中的表达模式。选取4个不同花色品种(橙色‘印丽’、粉色‘Grand Due’、红色‘安旺’、黄色‘洒金’)的花瓣,通过实时荧光定量PCR检测11个候选CiMYBs基因的表达模式,分析基因表达与花瓣花青素、类黄酮含量的关系;以拟南芥蛋白预测美人蕉中的蛋白互作。 结果 共鉴定出185个美人蕉R2R3-MYB基因,编码蛋白均为亲水性核定位蛋白,分布于9条染色体上。约94%的成员含有核心保守基序(Motif1、Motif2、Motif3),启动子区富含光响应元件。系统发育分析将其与拟南芥同源蛋白划分为30个亚组,其中11个基因与拟南芥中调控花色素合成的MYB蛋白同源;RNA-seq结果显示11个候选基因在不同花色美人蕉品种中差异表达;互作网络预测表明CiMYB65与花青素合成关键基因DFRA显著共表达,直接调控花青素合成。RT-qPCR分析发现CiMYB21、CiMYB39、CiMYB65和CiMYB20分别在‘Grand Due’‘洒金’‘安旺’‘印丽’中表达量最高,且基因表达量与花青素含量及黄酮醇含量呈显著正相关。 结论 美人蕉基因组含有185个R2R3-MYB基因,其中CiMYB20、CiMYB39、CiMYB65等基因可通过调节花青素合成参与调控美人蕉花色形成。
杨晨欣, 李梦秀, 姜唐, 张文娥, 潘学军. 美人蕉R2R3-MYB基因家族的鉴定及与花青素相关成员的表达分析[J]. 生物技术通报, 2025, 41(12): 201-213.
YANG Chen-xin, LI Meng-xiu, JIANG Tang, ZHANG Wen-e, PAN Xue-jun. Identification and Expression Analysis of Anthocyanin-associated R2R3-MYB Genes in Canna indica[J]. Biotechnology Bulletin, 2025, 41(12): 201-213.

图1 供试美人蕉材料表型A:转录组测序材料;B:基因组测序材料;C:RT-qPCR材料
Fig. 1 Phenotypes of tested Canna lily samplesA: RNA sequencing sample. B: Genomic DNA sample. C: RT-qPCR sample
基因名称 Gene name | 上游引物 Forward primer (5′-3′) | 下游引物 Reverse primer (5′-3′) |
|---|---|---|
| 18S | ATTCTATGGGTGGTGGTGC | CCATCCAATCGGTAGGAGC |
| CiMYB20 | AGCAGGGCTTTAACAAGGGG | GCTCCTCGGGCATCGATTTA |
| CiMYB39 | GAATGCAGGACTGCTGAGGT | GGTTGACGATGGTGTCGTCT |
| CiMYB65 | TGTTCCACCACCGCCATATC | CCAATCGGAGAACTCGCTGT |
| CiMYB116 | CGTCGTCAGTAGTACCGCTC | CTCCTCGTTGGGATCGACAG |
| CiMYB16 | GGCAGCCAACAAAAGGCTAC | GTCTCTGGATCTGCGTTGCT |
| CiMYB179 | GCTCATAGGCAACAGGTGGT | GAGCTGAGCTTTGACCGAGT |
| CiMYB8 | CCAGCAGCTCAAATCCTCCA | ACCAAGTCCTGCATGCTCTC |
| CiMYB80 | ACGGAGACTTGTTCGGTGTC | TGGCCATGTTGGGTTGTGAT |
| CiMYB21 | TTTGCTGACTTCAATGCGGC | CCTCCTCGATGAATCTCGCC |
| CiMYB171 | CGGAGGAAACTTACGGCACT | CTCCTCGGAGGCAATGAGTC |
| CiMYB68 | CCAGCCCTTGCTGTTACTGA | CCAGCAGCTCAAATCCTCCA |
表1 CiMYBs基因的引物
Table 1 Primers for CiMYBs genes
基因名称 Gene name | 上游引物 Forward primer (5′-3′) | 下游引物 Reverse primer (5′-3′) |
|---|---|---|
| 18S | ATTCTATGGGTGGTGGTGC | CCATCCAATCGGTAGGAGC |
| CiMYB20 | AGCAGGGCTTTAACAAGGGG | GCTCCTCGGGCATCGATTTA |
| CiMYB39 | GAATGCAGGACTGCTGAGGT | GGTTGACGATGGTGTCGTCT |
| CiMYB65 | TGTTCCACCACCGCCATATC | CCAATCGGAGAACTCGCTGT |
| CiMYB116 | CGTCGTCAGTAGTACCGCTC | CTCCTCGTTGGGATCGACAG |
| CiMYB16 | GGCAGCCAACAAAAGGCTAC | GTCTCTGGATCTGCGTTGCT |
| CiMYB179 | GCTCATAGGCAACAGGTGGT | GAGCTGAGCTTTGACCGAGT |
| CiMYB8 | CCAGCAGCTCAAATCCTCCA | ACCAAGTCCTGCATGCTCTC |
| CiMYB80 | ACGGAGACTTGTTCGGTGTC | TGGCCATGTTGGGTTGTGAT |
| CiMYB21 | TTTGCTGACTTCAATGCGGC | CCTCCTCGATGAATCTCGCC |
| CiMYB171 | CGGAGGAAACTTACGGCACT | CTCCTCGGAGGCAATGAGTC |
| CiMYB68 | CCAGCCCTTGCTGTTACTGA | CCAGCAGCTCAAATCCTCCA |
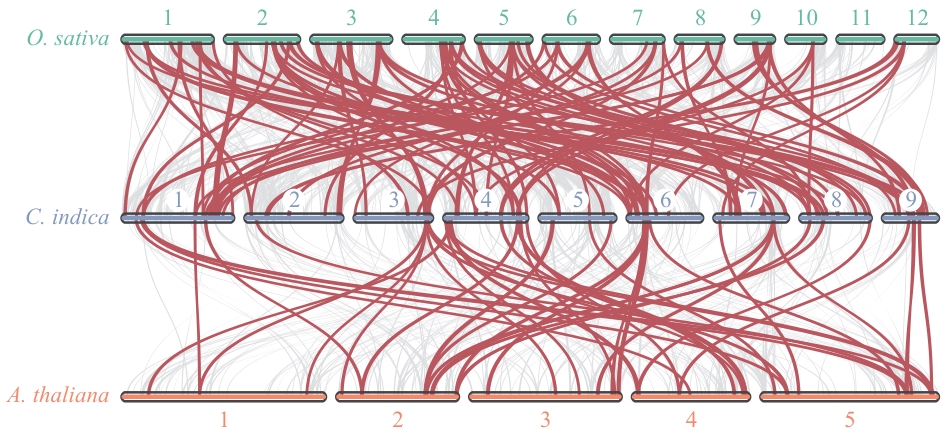
图5 美人蕉R2R3-MYB基因家族与拟南芥和水稻的蛋白序列共线性分析
Fig. 5 Collinearity analysis of protein sequences of the R2R3-MYB gene family in C. indica, A. thaliana,and O. sativa

图6 美人蕉R2R3-MYB基因结构分析A:Motif分析;B:基因结构域分析;C:基因结构分析
Fig. 6 Gene structures of R2R3-MYB genes in Canna lilyA: Motif analysis. B: Gene domain analysis. C: Gene structure analysis
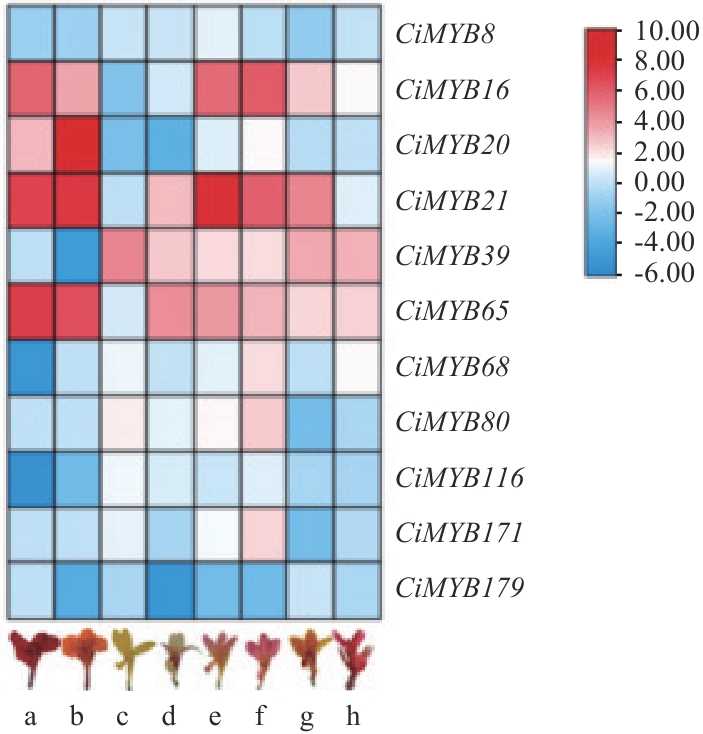
图8 部分美人蕉R2R3-MYB基因表达模式a:安旺;b:印丽;c:水生美人交后代1;d:水生美人蕉后代2;e:水生美人交后代3;f:水生美人蕉后代4;g:大花美人蕉后代1;h:大花美人蕉后代2
Fig. 8 Expression patterns of R2R3-MYB genes in Canna lilya: Anwang. b: Yinli. c: Progeny 1 of aquatic Canna. d: Progeny 2 of aquatic Canna. e: Progeny 3 of aquatic Canna. f: Progeny 4 of aquatic Canna. g: Progeny 1 of Canna generalis. h: Progeny 2 of Canna generalis

图9 花青素相关美人蕉R2R3-MYB与花青素合成结构基因的互作分析连接线表示基因间的共表达关系,其颜色编码反映互作性质,红色线条指示显著正相关(协同调控),蓝色线条代表显著负相关(抑制或拮抗作用),而线条粗细与共表达强度或统计置信度呈正相关
Fig. 9 Interaction analysis between anthocyanin-related Canna lilyThe connecting lines indicate co-expression relationships between genes, with color coding reflecting the nature of interactions. Red lines indicate significantly positive correlations (synergistic regulation), blue lines denote significantly negative correlations (repressive or antagonistic effects), and the thickness of the lines is positively correlated with the strength of co-expression or statistical confidence
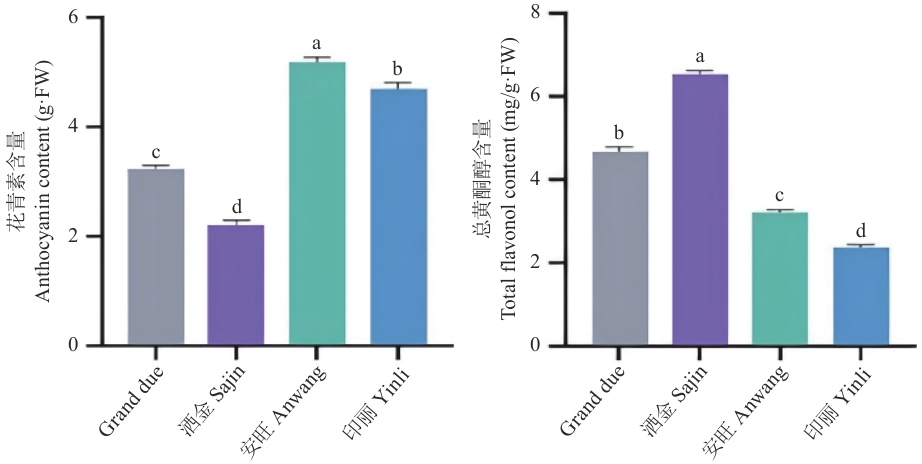
图10 美人蕉总花青素含量及总黄酮醇含量不同小写字母表示不同处理差异显著(P<0.05)。下同
Fig. 10 Contents of total anthocyanin and total flavonol in Canna lilyDifferent lowercase letters indicate significant differences among different treatments (P<0.05). The same below
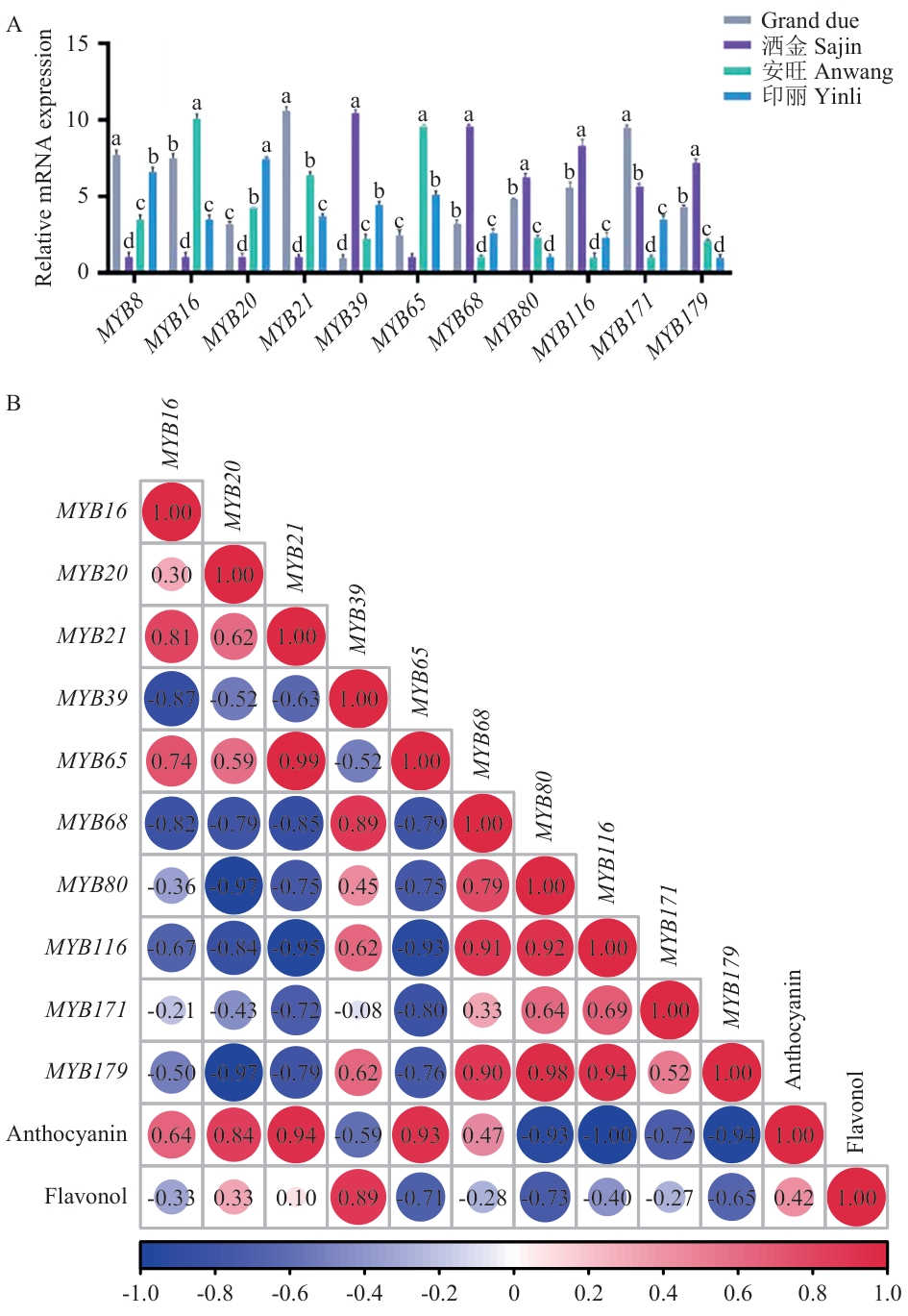
图11 候选R2R3-MYB基因在4个色系美人蕉品中的相对表达规律及相关性分析
Fig. 11 Relative expression patterns and correlation analysis of candidate R2R3-MYB genes in four cultivars of Canna lily
| [32] | Liu SA, Zhang HY, Meng ZL, et al. The LncNAT11-MYB11-F3'H/FLS module mediates flavonol biosynthesis to regulate salt stress tolerance in Ginkgo biloba [J]. J Exp Bot, 2025, 76(4): 1179-1201. |
| [33] | Li WF, Gao J, Ma ZH, et al. Molecular evolution and expression assessment of DFRs in apple [J]. Chem Biol Technol Agric, 2023, 10(1): 98. |
| [34] | Zhang L, Ma JN. Promoter cloning of VcCHS gene from blueberries and selection of its transcription factors [J]. Hortic Environ Biotechnol, 2025. DOI: https://doi.org/10.1007/s13580-025-00694-y . |
| [35] | Chen DZ, Wang CC, Liu Y, et al. Systematic identification of R2R3-MYB S6 subfamily genes in Brassicaceae and its role in anthocyanin biosynthesis in Brassica crops [J]. BMC Plant Biol, 2025, 25(1): 290. |
| [36] | Bhatia C, Gaddam SR, Pandey A, et al. COP1 mediates light-dependent regulation of flavonol biosynthesis through HY5 in Arabidopsis [J]. Plant Sci, 2021, 303: 110760. |
| [37] | Shi LY, Chen X, Wang K, et al. MrMYB6 from Chinese bayberry (Myrica rubra) negatively regulates anthocyanin and proanthocyanidin accumulation [J]. Front Plant Sci, 2021, 12: 685654. |
| [38] | Duan AQ, Tan SS, Deng YJ, et al. Genome-wide identification and evolution analysis of R2R3-MYB gene family reveals S6 subfamily R2R3-MYB transcription factors involved in anthocyanin biosynthesis in carrot [J]. Int J Mol Sci, 2022, 23(19): 11859. |
| [39] | Roy S, Tripathi AM, Yadav A, et al. Identification and expression analyses of miRNAs from two contrasting flower color cultivars of Canna by deep sequencing [J]. PLoS One, 2016, 11(1): e0147499. |
| [1] | Qi YT, Gu CH, Wang XJ, et al. Identification of the Eutrema salsugineum EsMYB90 gene important for anthocyanin biosynthesis [J]. BMC Plant Biol, 2020, 20(1): 186. |
| [2] | Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis [J]. Trends Plant Sci, 2010, 15(10): 573-581. |
| [3] | Nakatsuka T, Haruta KS, Pitaksutheepong C, et al. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers [J]. Plant Cell Physiol, 2008, 49(12): 1818-1829. |
| [4] | Yan HL, Pei XN, Zhang H, et al. MYB-mediated regulation of anthocyanin biosynthesis [J]. Int J Mol Sci, 2021, 22(6): 3103. |
| [5] | Poudel PR, Azuma A, Kobayashi S, et al. VvMYBAs induce expression of a series of anthocyanin biosynthetic pathway genes in red grapes (Vitis vinifera L.) [J]. Sci Hortic, 2021, 283: 110121. |
| [6] | Laitinen RAE, Ainasoja M, Broholm SK, et al. Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida [J]. J Exp Bot, 2008, 59(13): 3691-3703. |
| [7] | Yamagishi M, Shimoyamada Y, Nakatsuka T, et al. Two R2R3-MYB genes, homologs of Petunia AN2 regulate anthocyanin biosyntheses in flower Tepals, tepal spots and leaves of Asiatic hybrid lily [J]. Plant Cell Physiol, 2010, 51(3): 463-474. |
| [8] | Zheng YL, Chen YD, Liu ZG, et al. Important roles of key genes and transcription factors in flower color differences of Nicotiana alata [J]. Genes, 2021, 12(12): 1976. |
| [9] | 黄国涛, 欧阳底梅, 向其柏, 等. 美人蕉属品种分类研究 [J]. 南京林业大学学报: 自然科学版, 2005, 29(4): 20-24. |
| Huang GT, Ouyang DM, Xiang QB, et al. Studies on classification for cultivars of Canna L [J]. J Nanjing For Univ, 2005, 29(4): 20-24. | |
| [10] | Tripathi AM, Niranjan A, Roy S. Global gene expression and pigment analysis of two contrasting flower color cultivars of Canna [J]. Plant Physiol Biochem, 2018, 127: 1-10. |
| [11] | Liu MM, Li C, Jiang T, et al. Chromosome-scale genome assembly provides insights into flower coloration mechanisms of Canna indica [J]. Int J Biol Macromol, 2023, 251: 126148. |
| [12] | 肖伟军, 任小玉, 杨芯蕊, 等. 香蕉R 2R3-MYB基因家族鉴定及在果实发育中的表达分析 [J/OL]. 分子植物育种, 2024. . |
| Xiao WJ, Ren XY, Yang XR, et al. Genomic identification of the R 2R3-MYB gene family in banana and expression analysis during fruit development [J/OL]. Mol Plant Breed, 2024. . | |
| [13] | Song HY, Duan ZH, Wang Z, et al. Genome-wide identification, expression pattern and subcellular localization analysis of the JAZ gene family in Toona ciliata [J]. Ind Crops Prod, 2022, 178: 114582. |
| [14] | Chen GQ, He WZ, Guo XX, et al. Genome-wide identification, classification and expression analysis of the MYB transcription factor family in Petunia [J]. Int J Mol Sci, 2021, 22(9): 4838. |
| [15] | Tian SN, Ali MM, Ke ML, et al. Novel R2R3-MYB transcription factor LhMYB1 promotes anthocyanin accumulation in Lilium concolor var. pulchellum [J]. Horticulturae, 2024, 10(5): 509. |
| [16] | Stracke R, Ishihara H, Huep G, et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling [J]. Plant J, 2007, 50(4): 660-677. |
| [17] | Zhu ZP, Yu JX, Liu FF, et al. AeWRKY32 from okra regulates anthocyanin accumulation and cold tolerance in Arabidopsis [J]. J Plant Physiol, 2023, 287: 154062. |
| [18] | Chen LH, Hu B, Qin YH, et al. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors [J]. Plant Physiol Biochem, 2019, 136: 178-187. |
| [19] | 赵婷. 美人蕉属植物耐涝性评价及对淹水胁迫的形态生理与基因表达差异响应 [D]. 贵阳: 贵州大学, 2022. |
| Zhao T. Evaluation of waterlogging tolerance of Canna plants and its response to morphophysiological, physiological and gene expression differences under waterlogging stress [D]. Guiyang: Guizhou University, 2022. | |
| [20] | Shi YF, Lu TR, Lai SY, et al. Rosa rugosa R2R3-MYB transcription factors RrMYB12 and RrMYB111 regulate the accumulation of flavonols and anthocyanins [J]. Front Plant Sci, 2024, 15: 1477278. |
| [21] | Zhu J, Wang YZ, Zhou X, et al. Characterization of three novel R2R3-MYB transcription factors PrMYBi (1-3) repressing the anthocyanin biosynthesis in tree peony [J]. Hortic Plant J, 2024. DOI: https://doi.org/10.1016/j.hpj.2024.08.005 . |
| [22] | 刘雅芝, 邓惠敏, 彭嘉慧, 等. 铁十字秋海棠R2R3-MYB基因家族的鉴定及表达分析 [J]. 南方农业学报, 2024, 55(9): 2665-2678. |
| Liu YZ, Deng HM, Peng JH, et al. Identification and expression analysis of R2R3-MYB gene family in Begonia masoniana [J]. J South Agric, 2024, 55(9): 2665-2678. | |
| [23] | Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana [J]. Curr Opin Plant Biol, 2001, 4(5): 447-456. |
| [24] | Hu XM, Liang ZH, Sun TX, et al. The R2R3-MYB transcriptional repressor TgMYB4 negatively regulates anthocyanin biosynthesis in tulips (Tulipa gesneriana L.) [J]. Int J Mol Sci, 2024, 25(1): 563. |
| [25] | Alabd A, Ahmad M, Zhang X, et al. Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear [J]. Hortic Res, 2022, 9: uhac199. |
| [26] | Hsu CC, Chen YY, Tsai WC, et al. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp [J]. Plant Physiol, 2015, 168(1): 175-191. |
| [27] | Gu ZY, Zhu J, Hao Q, et al. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa) [J]. Plant Cell Physiol, 2019, 60(3): 599-611. |
| [28] | Wang CP, Li Y, Wang N, et al. An efficient CRISPR/Cas9 platform for targeted genome editing in rose (Rosa hybrida) [J]. J Integr Plant Biol, 2023, 65(4): 895-899. |
| [29] | Yang L, Wang G, Liu TT, et al. The MYB transcription factor RtAN2 promotes anthocyanin accumulation over proanthocyanidin during the fruit ripening of rose myrtle berries [J]. Ind Crops Prod, 2025, 224: 120433. |
| [30] | Cui C, Zhang K, Chai L, et al. Unraveling the mechanism of flower color variation in Brassica napus by integrated metabolome and transcriptome analyses [J]. Front Plant Sci, 2024, 15: 1419508. |
| [31] | Zuluaga DL, Gonzali S, Loreti E, et al. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants [J]. Funct Plant Biol, 2008, 35(7): 606-618. |
| [1] | 李锐, 胡婷, 陈树溦, 王尧, 王计平. 紫苏PfMYB80转录因子正向调控花青素的生物合成[J]. 生物技术通报, 2025, 41(6): 243-255. |
| [2] | 郭涛, 艾丽皎, 邹世慧, 周玲, 李学梅. 山茶CjRAV1调控开花延迟的功能研究[J]. 生物技术通报, 2025, 41(6): 208-217. |
| [3] | 周熠, 刘勇波. 基因组进化过程中的基因丢失机制与功能研究进展[J]. 生物技术通报, 2025, 41(6): 38-48. |
| [4] | 刘红利, 马一丹, 王婉茹, 杨娅茹, 贺丹, 刘艺平, 孔德政. 荷花NnCYP707A1的克隆及功能分析[J]. 生物技术通报, 2025, 41(5): 197-207. |
| [5] | 刘源, 赵冉, 卢振芳, 李瑞丽. 植物类胡萝卜素生物代谢途径及其功能研究进展[J]. 生物技术通报, 2025, 41(5): 23-31. |
| [6] | 田琴, 刘奎, 吴翔纬, 纪媛媛, 曹一博, 张凌云. 转录因子VcMYB17调控蓝莓抗旱性的功能研究[J]. 生物技术通报, 2025, 41(4): 198-210. |
| [7] | 杨朝结, 张兰, 陈红, 黄娟, 石桃雄, 朱丽伟, 陈庆富, 李洪有, 邓娇. 苦荞转录因子基因FtbHLH3调控类黄酮生物合成的功能鉴定[J]. 生物技术通报, 2025, 41(4): 134-144. |
| [8] | 李斌, 苏香萍, 刘畅, 王玉兵, 张勇洪, 周超, 徐青. 玄参科植物叶绿体基因组特征及系统发育分析[J]. 生物技术通报, 2025, 41(3): 240-254. |
| [9] | 苗昊翔, 张颖, 郭世鹏, 张健. 一株高产γ-氨基丁酸短乳杆菌TCCC13007全基因组测序及比较基因组分析[J]. 生物技术通报, 2025, 41(11): 166-176. |
| [10] | 刘梓琦, 钟沛, 李琴, 郭成, 张艳梅, 张乃锋, 屠焰, 刁其玉, 毕研亮. CRISPR/Cas9技术在益生菌编辑中的应用与进展[J]. 生物技术通报, 2025, 41(11): 89-99. |
| [11] | 秦文俊, 熊言杰, 赵冉, 马萧然, 叶霄萌, 宋江华. 甘蓝ARF基因家族的鉴定及其在非生物胁迫下的表达分析[J]. 生物技术通报, 2025, 41(10): 253-263. |
| [12] | 杜品廷, 吴国江, 王振国, 李岩, 周伟, 周亚星. 高粱CPP基因家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 132-142. |
| [13] | 李雨晴, 吴楠, 罗建让. 卵叶牡丹花色苷合成相关基因bHLH的克隆与功能分析[J]. 生物技术通报, 2024, 40(8): 174-185. |
| [14] | 聂祝欣, 郭瑾, 乔子洋, 李微薇, 张学燕, 刘春阳, 王静. 黑果枸杞不同发育时期果实花色苷合成的转录组分析[J]. 生物技术通报, 2024, 40(8): 106-117. |
| [15] | 周麟, 黄顺满, 苏文坤, 姚响, 屈燕. 滇山茶bHLH基因家族鉴定及花色形成相关基因筛选[J]. 生物技术通报, 2024, 40(8): 142-151. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||