








生物技术通报 ›› 2021, Vol. 37 ›› Issue (5): 98-107.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1149
白福美1( ), 李至敏2, 王小琴1, 胡紫微1, 鲍玲玲1, 李志敏1,3(
), 李至敏2, 王小琴1, 胡紫微1, 鲍玲玲1, 李志敏1,3( )
)
收稿日期:2020-09-09
出版日期:2021-05-26
发布日期:2021-06-11
作者简介:白福美,女,硕士研究生,研究方向:酶化学;E-mail: 基金资助:
BAI Fu-mei1( ), LI Zhi-min2, WANG Xiao-qin1, HU Zi-wei1, BAO Ling-ling1, LI Zhi-min1,3(
), LI Zhi-min2, WANG Xiao-qin1, HU Zi-wei1, BAO Ling-ling1, LI Zhi-min1,3( )
)
Received:2020-09-09
Published:2021-05-26
Online:2021-06-11
摘要:
旨在对集胞藻PCC6803中slr1022基因编码的N-乙酰鸟氨酸转氨酶进行生化表征及结构分析,为进一步研究该酶的催化功能及机制奠定理论基础。以集胞藻PCC6803基因组为模板,通过PCR扩增获得slr1022基因,将其连接到表达载体pET-28a上,转化大肠杆菌BL21(DE3)感受态。经IPTG诱导,Ni-NTA亲和层析纯化后获得重组Slr1022蛋白,然后通过紫外分光光度法对Slr1022蛋白的催化功能进行表征,并运用生物信息学软件对该蛋白进行结构分析。成功构建pET28a-slr1022重组表达质粒,并诱导表达重组Slr1022蛋白,SDS-PAGE电泳鉴定该蛋白分子量约为50 kD,与理论大小相符。Slr1022蛋白与底物N-乙酰鸟氨酸的结合常数Km和最大反应速度Vmax分别是0.12 mmol/L和0.60 μmol/(L·s),并且Slr1022蛋白与另一底物α-酮戊二酸的Km和Vmax分别是0.039 mmol/L和0.65 μmol/(L·s)。Slr1022蛋白在pH 8.5时催化活性最强。Slr1022蛋白和其他来源的N-乙酰鸟氨酸转氨酶氨基酸序列有一定的同源性,而且活性位点的氨基酸残基高度保守。成功克隆并表达纯化出Slr1022蛋白,酶学性质和生物信息学研究表明Slr1022蛋白为N-乙酰鸟氨酸转氨酶。
白福美, 李至敏, 王小琴, 胡紫微, 鲍玲玲, 李志敏. 集胞藻PCC6803中N-乙酰鸟氨酸转氨酶的生化表征及结构分析[J]. 生物技术通报, 2021, 37(5): 98-107.
BAI Fu-mei, LI Zhi-min, WANG Xiao-qin, HU Zi-wei, BAO Ling-ling, LI Zhi-min. Biochemical Characterization and Structural Analysis of N-acetylornithine Transaminase from Synechocystis sp. PCC6803[J]. Biotechnology Bulletin, 2021, 37(5): 98-107.
| Primer | Primer sequence(5'-3') | Restriction enzyme |
|---|---|---|
| slr1022-F | GGAATTCCATATGACCTATTCC-CCTGTTGTTGAATC | Nde I |
| slr1022-R | CCGCTCGAGTCAAACCAAAGT-GGCGATCGCCTGAC | Xho I |
表1 PCR引物序列
Table 1 PCR primer sequences
| Primer | Primer sequence(5'-3') | Restriction enzyme |
|---|---|---|
| slr1022-F | GGAATTCCATATGACCTATTCC-CCTGTTGTTGAATC | Nde I |
| slr1022-R | CCGCTCGAGTCAAACCAAAGT-GGCGATCGCCTGAC | Xho I |
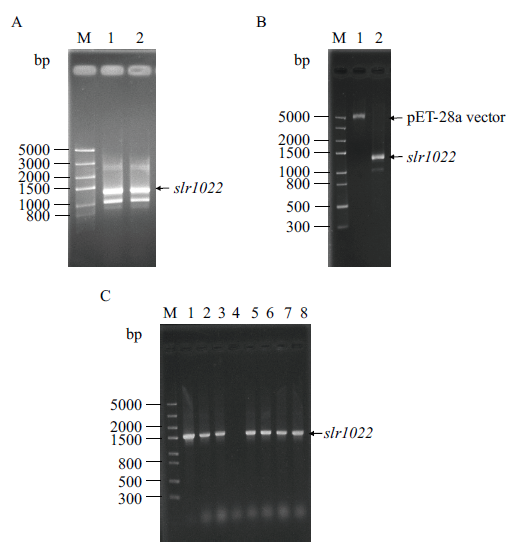
图1 集胞藻PCC6803中slr1022基因克隆及pET28a-slr1022重组质粒的构建 A:slr1022基因PCR扩增产物。M:DNA分子标准;1-2:PCR扩增产物。B:slr1022基因和pET-28a载体双酶切产物。M:DNA分子标准;1-2:分别为pET-28a载体和slr1022基因双酶切产物。C:pET28a-slr1022重组质粒鉴定。M:DNA分子标准;1:正对照;2-8:菌落PCR产物
Fig. 1 Cloning of slr1022 gene from Synechocystis sp. PCC6803 and construction of its recombinant plasmid pET28a-slr1022 A: PCR amplification product of slr1022 gene. M: DNA marker. Lane 1-2: PCR product. B: Double enzymatic digestion products of slr1022 gene and pET-28a vector. M: DNA marker. Lane 1-2: Double enzymatic digestion product of pET-28a vector and slr1022 gene, respectively. C: Identification of recombinant plasmid pET28a-slr1022. M: DNA marker. Lane 1: Positive control. Lane 2-8: Colony PCR product

图2 重组Slr1022蛋白的诱导表达 M:蛋白质标准;1-3:pET-28a空载体的表达(分别为细胞破碎液,离心后上清液和离心后沉淀重悬液);4-6:pET28a-slr1022重组质粒的表达(分别为细胞破碎液,离心后上清液和离心后沉淀重悬液)
Fig. 2 Expression of recombinant Slr1022 protein M: Protein marker. Lane 1-3: Expression of pET-28a plasmid (cell lysate, supernatant and pellet, respectively). Lane 4-6: Expression of pET28a-slr1022 recombinant plasmid (cell lysate, supernatant and pellet, respectively)
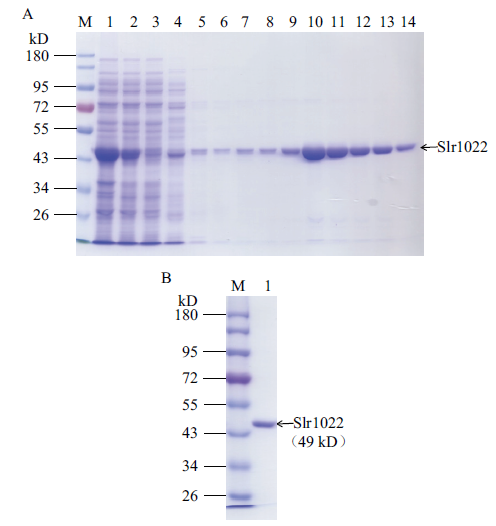
图3 重组Slr1022蛋白的亲和层析纯化结果电泳图 A:重组Slr1022蛋白的纯化。M:蛋白质标准;1-3:分别为细胞破碎液,离心后上清液和流穿液;4-8:分别为20、40、60、80和100 mmol/L 咪唑洗脱液;9-14:200 mmol/L 咪唑洗脱液。B:纯化后的Slr1022蛋白。M:蛋白质标准;1:纯化后的Slr1022蛋白
Fig. 3 SDS-PAGE of recombinant Slr1022 protein A: Purification of recombinant Slr1022 protein. M: Protein marker. Lane 1-3: Cell lysate, supernatant and flow through. Lane 4-8: 20, 40, 60, 80, 100 mmol/L imidazole elution. 9-14: 200 mmol/L imidazole elution. B: The purified Slr1022 protein. M: Protein marker. Lane 1: The purified Slr1022 protein
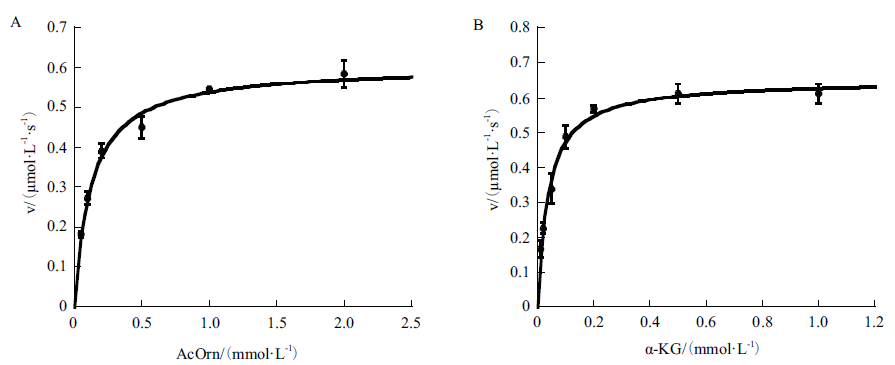
图4 重组Slr1022蛋白的酶动力学参数 A:初始反应速度随N-乙酰鸟氨酸浓度的变化曲线,固定α-酮戊二酸浓度为1 mmol/L;B:初始反应速度随α-酮戊二酸浓度的变化曲线,固定N-乙酰鸟氨酸浓度为2 mmol/L。数据以平均值 ± 标准差表示,n=3
Fig. 4 Kinetic profiles of recombinant Slr1022 protein A: Plot of the initial velocities as function of N-acetylornithine concentrations, the concentration of α-ketoglutarate was fixed at 1 mmol/L. B: Plot of the initial velocities as function of α-ketoglutarate concentrations, the concentration of N-acetylornithine was fixed at 2 mmol/L. Data are expressed as the mean ± standard deviation, n = 3

图5 pH对重组Slr1022蛋白催化活性的影响 数据以平均值 ± 标准差表示,n=3
Fig. 5 Effects of pH on the catalytic activity of recombinant Slr1022 protein Data are expressed as the mean ± standard deviation, n = 3

图6 不同来源的N-乙酰鸟氨酸转氨酶的氨基酸序列比对 slr1022、2E54、6W7X、2EH6和2PB0分别为来源于集胞藻PCC6803、海栖热袍菌、嗜麦芽窄单胞菌、超嗜热菌和鼠伤寒沙门氏菌的N-乙酰鸟氨酸转氨酶。▲表示与PLP结合的氨基酸残基;★表示与PLP形成希夫碱的氨基酸残基;◆表示来自另一单体和PLP结合的氨基酸残基
Fig. 6 Protein sequences alignment of N-acetylornithine aminotransferases from different sources slr1022, 2E54, 6W7X, 2EH6 and 2PB0 were N-acetylornithine aminotransferases from Synechocystis PCC6803, Thermotoga maritima, Stenotrophomonas maltophilia, Aquifex aeolicus and Salmonella typhimurium respectively. Residues interacting with PLP are marked by▲, and★ indicates the residues forming Schiff base with PLP and ◆ indicates the residues from the other subunit which interact with PLP

图7 Slr1022蛋白分子同源建模结构 A:Slr1022蛋白的卡通示意图。B:活性中心放大示意图。C:活性中心保守氨基酸残基。α-螺旋、β-折叠及无规则卷曲的颜色分别为红色、黄色和绿色。辅因子PLP与保守氨基酸残基为棍棒结构,其碳原子颜色分别为浅蓝色与绿色。黄色虚线为与PLP相互作用的氢键。该模拟结构由SWISS-MODEL软件基于海栖热袍菌中N-乙酰鸟氨酸转氨酶晶体结构(PDB ID:2E54)绘制
Fig. 7 Homologous modeling structure of Slr1022 protein A: Cartoon structure of Slr1022 protein. B: Enlarged schematic diagram of active site. C: The conserved amino acid residues in the active site. The colors of α-helix, β-strands and loops are depicted as red, yellow and green, respectively. The cofactor PLP and the conserved amino acid residues are shown as sticks, and their carbon atoms are light blue and green, respectively. The yellow dotted lines indicate the hydrogen bonds interacting with PLP. The modeled structure is depicted by SWISS-MODEL based on the structure of N-acetylornithine aminotransferase of Thermotoga maritima (PBD: 2E54)
| [1] | Burkill PH, Leakey RJG, Owens NJP, et al. Synechococcus and its importance to the microbial foodweb of the northwestern Indian Ocean[J]. Deep-Sea Research II, 1993,40(3):773-782. |
| [2] | 裴广胜. 模式蓝细菌集胞藻中分子调控系统的解析[D]. 天津:天津大学, 2017. |
| Pei GS. Functions analysis of regulatory molecules in the model cyanobacterium Synechocystis sp. PCC 6803[D]. Tianjin:Tianjin University, 2017. | |
| [3] | Stanier RY, Kunisawa R, Mandel R, et al. Purification and properties of unicellular blue-green algae(order chroococcales)[J]. Bacterological Reviews, 1971,35(2):171-205. |
| [4] |
Kaneko T, Sato S, Kotani H, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. sequence determination of the entire genome and assignment of potential protein-coding regions(Supplement)[J]. DNA Research, 1996,3(3):185-209.
pmid: 8905238 |
| [5] | Gao Z, Zhao H, Li Z, et al. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria[J]. Energy & Environmental Science, 2012,5(12):9857-9865. |
| [6] |
Aizouq M, Peisker H, Gutbrod K, et al. Triacylglycerol and phytyl ester synjournal in Synechocystis sp. PCC6803[J]. PNAS, 2020,117(11):6216-6222.
doi: 10.1073/pnas.1915930117 URL |
| [7] | 张兵, 王利红, 徐玉新, 等. 集胞藻(Synechocystis sp. PCC6803)对砷吸收转化特性的初步研究[J]. 生态毒理学报, 2011,6(6):629-633. |
| Zhang B, Wang LH, Xu YX, et al. Study on absorption and transformation of arsenic in blue alga(Synechocystis sp. PCC6803)[J]. Asian J Ecotox, 2011,6(6):629-633. | |
| [8] |
Pearce J, Carr NG. The metabolism of acetate by the blue-green algae, Anabaena variabilis and Anacystis nidulans[J]. Journal of General Microbiology, 1967,49:301-313.
pmid: 6077932 |
| [9] |
Xiong W, Brune D, Vermaas WF. The γ-aminobutyric acid shunt contributes to closing the tricarboxylic acid cycle in Synechocystis sp. PCC 6803[J]. Mol Microbiol, 2014,93(4):786-796.
doi: 10.1111/mmi.12699 URL |
| [10] | Zhang S, Qian X, Chang S, et al. Natural and synthetic variants of the tricarboxylic acid cycle in cyanobacteria:introduction of the GABA shunt into Synechococcus sp. PCC 7002[J]. Frontiers in Microbiology, 2016,7:1972. |
| [11] |
Philipp C, Mehta PK. From cofactor to enzymes. The molecular evolution of pyridoxal-5’-phosphate-dependent enzymes[J]. The Chemical Record, 2001,1:436-447.
doi: 10.1002/(ISSN)1528-0691 URL |
| [12] |
Rajaram V, Ratna Prasuna P, Savithri HS, et al. Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium:studies on substrate specificity and inhibitor binding[J]. Proteins, 2008,70(2):429-441.
doi: 10.1002/prot.21567 URL |
| [13] |
Ledwidge R, Blanchard JS. The dual biosynthetic capability of N-acetylornithine aminotransferase in arginine and lysine biosynjournal[J]. Biochemistry, 1999,38(10):3019-3024.
doi: 10.1021/bi982574a URL |
| [14] | 王丹. 牛肝谷氨酸脱氢酶的分离纯化及酶学性质和功能基团研究[D]. 重庆:西南大学, 2016. |
| Wang D. Isolation, purification, properties and modification of groups of the glutamate dehydrogenase from bovine liver[D]. Chongqing:Southwest University, 2016. | |
| [15] | 徐美娟, 张显, 饶志明, 等. 钝齿棒杆菌N-乙酰鸟氨酸转氨酶的克隆表达分析及其重组菌的精氨酸发酵[J]. 生物工程学报, 2011,27(7):1013-1023. |
| Xu M, Zhang X, Rao Z, et al. Cloning, expression and characterization of N-acetylornithine aminotransferase from Corynebacterium crenatum and its effects on L-arginine fermentation[J]. Chinese J Biotechnol, 2011,27(7):1013-1023. | |
| [16] |
Friedrich B, Friedrich CG, Magasanik B. Catabolic N2-acetylornithine 5-aminotransferase of Klebsiella aerogenes:control of synjournal by induction, catabolite repression, and activation by glutamine synthetase[J]. J Bacteriol, 1978,133(2):686-691.
pmid: 24039 |
| [17] |
Mehta PK, Hale TI, Christen P. Aminotransferases:demonstration of homology and division into evolutionary subgroups[J]. European Journal of Biochemistry 1993,214:549-561.
pmid: 8513804 |
| [18] |
Shen BW, Hennig M, Hohenester E, et al. Crystal structure of human recombinant ornithine aminotransferase[J]. Journal of Molecular Biology, 1998,277:81-102.
doi: 10.1006/jmbi.1997.1583 URL |
| [19] |
Toney MD, Pascarella S, Biase DD. Active site model for γ-aminobutyrate aminotransferase explains substrate specificity and inhibitor reactivities[J]. Protein Science, 1995,4:2366-2374.
pmid: 8563634 |
| [20] |
Steinhauser D, Fernie AR, Araujo WL. Unusual cyanobacterial TCA cycles:not broken just different[J]. Trends in Plant Science, 2012,17(9):503-509.
doi: 10.1016/j.tplants.2012.05.005 pmid: 22658681 |
| [21] |
Fait A, Nesi AN, et al. Targeted enhancement of glutamate-to-gamma-aminobutyrate conversion in Arabidopsis seeds affects carbon-nitrogen balance and storage reserves in a development-dependent manner[J]. Plant Physiol, 2011,157:1026-1042.
doi: 10.1104/pp.111.179986 URL |
| [22] |
Bouche N, Fait A, Bouchez D, et al. Mitochondrial succinic-semialdehyde dehydrogenase of the γ-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants[J]. PNAS, 2003,100(11):6843-6848.
doi: 10.1073/pnas.1037532100 URL |
| [23] |
Palanivelu R, Edlund AF, Brass L, et al. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels[J]. Cell, 2003,114:47-59.
doi: 10.1016/S0092-8674(03)00479-3 URL |
| [24] | Decavel C, Van Den Pol AN. GABA:a dominant neurotransmitter in the hypothalamus[J]. The Journal of Comparam Neurologyneurology, 1990,302:1019-1037. |
| [25] |
Granger AJ, Wallace ML, Sabatini BL. Multi-transmitter neurons in the mammalian central nervous system[J]. Current Opinion in Neurobiology, 2017,45:85-91.
doi: S0959-4388(17)30016-8 pmid: 28500992 |
| [1] | 梅欢, 李玥, 刘可蒙, 刘吉华. 小檗碱桥酶高效原核表达及生物合成l-SLR的研究[J]. 生物技术通报, 2023, 39(7): 277-287. |
| [2] | 索青青, 吴楠, 杨慧, 李莉, 王锡锋. 水稻咖啡酰辅酶A-O-甲基转移酶基因的原核表达、抗体制备和应用[J]. 生物技术通报, 2022, 38(8): 135-141. |
| [3] | 覃雪晶, 王雨涵, 曹一博, 张凌云. 青杄PwHAP5基因原核表达及多克隆抗体制备[J]. 生物技术通报, 2022, 38(8): 142-149. |
| [4] | 王光丽, 范婵, 王辉, 卢惠芳, 夏灵尹, 黄健, 闵迅. 霍乱弧菌溶血素HlyA的原核表达、纯化及多克隆抗体制备与鉴定[J]. 生物技术通报, 2022, 38(7): 269-277. |
| [5] | 汪巧菊, 胡雨萌, 温亚亚, 宋丽, 孟闯, 潘志明, 焦新安. 新型冠状病毒S1蛋白的表达及活性鉴定[J]. 生物技术通报, 2022, 38(3): 157-163. |
| [6] | 沈俊强, 张莉萍, 于瑞明, 王永录, 潘丽, 刘霞, 刘新生. 猪嵴病毒结构蛋白VP0与VP1原核表达及间接ELISA方法的建立[J]. 生物技术通报, 2022, 38(10): 243-253. |
| [7] | 山草梅, 叶蕾, 张连虎, 况卫刚, 孙晓棠, 马建, 崔汝强. 水稻抗潜根线虫基因OsRAI1的克隆及功能分析[J]. 生物技术通报, 2021, 37(7): 146-155. |
| [8] | 曾福源, 苏泽辉, 周诗慧, 谢妙, 庞欢瑛. 溶藻弧菌PEPCK蛋白原核表达及其乙酰化、琥珀酰化修饰的鉴定[J]. 生物技术通报, 2021, 37(5): 84-91. |
| [9] | 张西西, 张怡青, 李玉林, 韩笑, 王国强, 王晓军, 王旭东, 王云龙. 新型冠状病毒(SARS-CoV-2)N蛋白C端重组蛋白的原核表达、纯化及应用[J]. 生物技术通报, 2021, 37(5): 92-97. |
| [10] | 瞿欢, 李成, 陈汭, 廖艺杰, 曹三杰, 文翼平, 颜其贵, 黄小波. 猪δ冠状病毒S1-CTD的截短表达及间接ELISA抗体方法的建立[J]. 生物技术通报, 2021, 37(5): 273-280. |
| [11] | 彭利忠, 张鹏, 周雯雯, 曾旭辉, 张小宁. 精子特异性蛋白Cabs1多克隆抗体的制备及多用途验证[J]. 生物技术通报, 2021, 37(3): 261-270. |
| [12] | 贺扬, 余巧玲, 王均, 覃川杰, 李华涛. 罗非鱼原核表达基因研究进展[J]. 生物技术通报, 2021, 37(2): 195-202. |
| [13] | 唐禄, 董丽平, 尹茉莉, 刘磊, 董媛, 王会岩. 成纤维细胞生长因子20单克隆抗体的制备及鉴定[J]. 生物技术通报, 2021, 37(10): 179-185. |
| [14] | 段应策, 胡姿仪, 杨帆, 李金涛, 邬向丽, 张瑞颖. 香菇草酰乙酸水解酶基因LeOAH1克隆及表达分析[J]. 生物技术通报, 2020, 36(9): 227-234. |
| [15] | 陈汭, 付嘉钰, 刘浩宇, 李成, 赵玉佳, 胡靖飞, 瞿欢, 曹三杰, 文心田, 文翼平, 赵勤, 伍锐, 黄小波. 猪δ冠状病毒(PDCoV)N蛋白的原核表达及多克隆抗体制备[J]. 生物技术通报, 2020, 36(8): 104-110. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||