








生物技术通报 ›› 2023, Vol. 39 ›› Issue (1): 115-126.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0378
周家燕1( ), 邹建1, 陈卫英1, 吴一超1, 陈奚潼1, 王倩1, 曾文静1, 胡楠2(
), 邹建1, 陈卫英1, 吴一超1, 陈奚潼1, 王倩1, 曾文静1, 胡楠2( ), 杨军1(
), 杨军1( )
)
收稿日期:2022-03-29
出版日期:2023-01-26
发布日期:2023-02-02
作者简介:周家燕,女,硕士研究生,研究方向:植物遗传与发育;E-mail: 基金资助:
ZHOU Jia-yan1( ), ZOU Jian1, CHEN Wei-ying1, WU Yi-chao1, CHEN Xi-tong1, WANG Qian1, ZENG Wen-jing1, HU Nan2(
), ZOU Jian1, CHEN Wei-ying1, WU Yi-chao1, CHEN Xi-tong1, WANG Qian1, ZENG Wen-jing1, HU Nan2( ), YANG Jun1(
), YANG Jun1( )
)
Received:2022-03-29
Published:2023-01-26
Online:2023-02-02
摘要:
多数重要的功能基因属于多基因家族,这些家族成员间存在功能冗余,高效的多基因干扰体系对研究多基因家族成员的生物学功能及其分子调控机制具有重要意义。对pCAMBIA1301载体改造,构建了适用于植物的多基因干扰体系pCAMBIA1301m和pCAMBIA1301s。使用该多基因干扰体系构建了四基因的干扰载体pCAMBIA1301m:35S∷SlPP2C1-2-3-4,4个目标基因为来源于番茄PP2C家族A组的PP2C1、PP2C2、PP2C3和PP2C4,并通过遗传转化导入番茄,用GUS染色和PCR检测转基因阳性植株,再利用RT-qPCR技术检测T1和T2代转基因植株中目标基因的干扰效率,用T2代种子分析转基因番茄对ABA敏感性。结果表明,应用该干扰体系成功获得了四基因干扰的转基因植株35S∷SlPP2C1-2-3-4。在转基因番茄中4个目标基因的表达量显著低于野生型,其干扰效率均高于70%,转基因番茄种子萌发具有强烈的ABA不敏感性。多基因干扰体系能高效地同时沉默多个目标基因。
周家燕, 邹建, 陈卫英, 吴一超, 陈奚潼, 王倩, 曾文静, 胡楠, 杨军. 植物多基因干扰载体体系构建与效用分析[J]. 生物技术通报, 2023, 39(1): 115-126.
ZHOU Jia-yan, ZOU Jian, CHEN Wei-ying, WU Yi-chao, CHEN Xi-tong, WANG Qian, ZENG Wen-jing, HU Nan, YANG Jun. Construction of Multi-gene Interference System for Plant and Analysis of Its Application Efficiency[J]. Biotechnology Bulletin, 2023, 39(1): 115-126.
| 基因名称 Gene name | 番茄数据库基因ID Gene ID in tomato genome database |
|---|---|
| SlPP2C1 | Solyc03g121880.2.1 |
| SlPP2C2 | Solyc12g096020.1.1 |
| SlPP2C3 SlPP2C4 | Solyc08g062650.2.1 Solyc07g040990.2.1 |
表1 本文分析的4个PP2C基因的相关信息
Table 1 Information of four PP2C genes used in this paper
| 基因名称 Gene name | 番茄数据库基因ID Gene ID in tomato genome database |
|---|---|
| SlPP2C1 | Solyc03g121880.2.1 |
| SlPP2C2 | Solyc12g096020.1.1 |
| SlPP2C3 SlPP2C4 | Solyc08g062650.2.1 Solyc07g040990.2.1 |
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| SlPP2C1-F正 | GCGAGCTCGTTGATGTTGGTAGAGTTCCCTTG | 干扰片段扩增引物 Primers for amplifying interference fragment |
| SlPP2C1-R-B | TCCGAATCAGCCTTTCTATCTAAACTCCTTTCTTCC | |
| SlPP2C2-F-B | GATAGAAAGGCTGATTCGGAGAGTGACCTTAG | |
| SlPP2C2-R正 | ACGCGTCGACGAGAATCCACGAAATCCTGACC | |
| SlPP2C1-F反 | CTCTAGAGTTGATGTTGGTAGAGTTCCCTTG | |
| SlPP2C1-R-B | TCCGAATCAGCCTTTCTATCTAAACTCCTTTCTTCC | |
| SlPP2C2-F-B | GATAGAAAGGCTGATTCGGAGAGTGACCTTAG | |
| SlPP2C2-R反 | TCCCCCGGGGAGAATCCACGAAATCCTGACC | |
| SlPP2C3-F正 | GCGAGCTCATGTGTTTGGCGGTTGCTTTGC | |
| SlPP2C3-R-B | CTATGATTCGGCATCTACCAAATTCTGTCCGTC | |
| SlPP2C4-F-B | TGGTAGATGCCGAATCATAGAGTGTCCAGGG | |
| SlPP2C4-R正 | ACGCGTCGACTTAGCCAAACTACACGAAAAAGAC | |
| SlPP2C3-F反 | CTCTAGAATGTGTTTGGCGGTTGCTTTGC | |
| SlPP2C3-R-B | CTATGATTCGGCATCTACCAAATTCTGTCCGTC | |
| SlPP2C4-F-B | TGGTAGATGCCGAATCATAGAGTGTCCAGGG | |
| SlPP2C4-R反 | TCCCCCGGGTTAGCCAAACTACACGAAAAAGAC | |
| qSlPP2C1-F | TAGCTGCACCTCTGAGCCTA | RT-qPCR引物Primers for RT-qPCR |
| qSlPP2C1-R | CTGCTTCTTTGCCACGATAA | |
| qSlPP2C2-F | CAGCAGAATGCTTGTCGAAT | |
| qSlPP2C2-R | ATGAGGCCAATTGTGTTGAA | |
| qSlPP2C3-F | GTCGCCATTGTTTGTTCATC | |
| qSlPP2C3-R | TCTTCTCGATTTGGCTTGTG | |
| qSlPP2C4-F | GATGGGCTATGGGATGTCTT | |
| qSlPP2C4-R | CTTGAGCAGCAGGATCTACG | |
| qSlUBI-F | GCCGACTACAACATCCAGAAGG | RT-qPCR内参引物Reference primers for RT-qPCR |
| qSlUBI-R | TGCAACACAGCGAGCTTAACC | |
| Loop-JC-F | ACAAGTTCAGCGTGTCCGGCGA | 载体元件特异引物Special primers for vector element |
| Loop-JC-R | TCGCCGGACACGCTGAACTTGT |
表2 本文使用的所有引物
Table 2 All primers used in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| SlPP2C1-F正 | GCGAGCTCGTTGATGTTGGTAGAGTTCCCTTG | 干扰片段扩增引物 Primers for amplifying interference fragment |
| SlPP2C1-R-B | TCCGAATCAGCCTTTCTATCTAAACTCCTTTCTTCC | |
| SlPP2C2-F-B | GATAGAAAGGCTGATTCGGAGAGTGACCTTAG | |
| SlPP2C2-R正 | ACGCGTCGACGAGAATCCACGAAATCCTGACC | |
| SlPP2C1-F反 | CTCTAGAGTTGATGTTGGTAGAGTTCCCTTG | |
| SlPP2C1-R-B | TCCGAATCAGCCTTTCTATCTAAACTCCTTTCTTCC | |
| SlPP2C2-F-B | GATAGAAAGGCTGATTCGGAGAGTGACCTTAG | |
| SlPP2C2-R反 | TCCCCCGGGGAGAATCCACGAAATCCTGACC | |
| SlPP2C3-F正 | GCGAGCTCATGTGTTTGGCGGTTGCTTTGC | |
| SlPP2C3-R-B | CTATGATTCGGCATCTACCAAATTCTGTCCGTC | |
| SlPP2C4-F-B | TGGTAGATGCCGAATCATAGAGTGTCCAGGG | |
| SlPP2C4-R正 | ACGCGTCGACTTAGCCAAACTACACGAAAAAGAC | |
| SlPP2C3-F反 | CTCTAGAATGTGTTTGGCGGTTGCTTTGC | |
| SlPP2C3-R-B | CTATGATTCGGCATCTACCAAATTCTGTCCGTC | |
| SlPP2C4-F-B | TGGTAGATGCCGAATCATAGAGTGTCCAGGG | |
| SlPP2C4-R反 | TCCCCCGGGTTAGCCAAACTACACGAAAAAGAC | |
| qSlPP2C1-F | TAGCTGCACCTCTGAGCCTA | RT-qPCR引物Primers for RT-qPCR |
| qSlPP2C1-R | CTGCTTCTTTGCCACGATAA | |
| qSlPP2C2-F | CAGCAGAATGCTTGTCGAAT | |
| qSlPP2C2-R | ATGAGGCCAATTGTGTTGAA | |
| qSlPP2C3-F | GTCGCCATTGTTTGTTCATC | |
| qSlPP2C3-R | TCTTCTCGATTTGGCTTGTG | |
| qSlPP2C4-F | GATGGGCTATGGGATGTCTT | |
| qSlPP2C4-R | CTTGAGCAGCAGGATCTACG | |
| qSlUBI-F | GCCGACTACAACATCCAGAAGG | RT-qPCR内参引物Reference primers for RT-qPCR |
| qSlUBI-R | TGCAACACAGCGAGCTTAACC | |
| Loop-JC-F | ACAAGTTCAGCGTGTCCGGCGA | 载体元件特异引物Special primers for vector element |
| Loop-JC-R | TCGCCGGACACGCTGAACTTGT |

图1 多基因沉默载体系统 A:原始载体系统pCAMBIA1301;B:改进后的载体系统pCAMBIA1301m;C:改进后的载体系统pCAMBIA1301s;D:多基因干扰载体体系
Fig. 1 Multi-genes silencing system A:Original vector system pCAMBIA1301. B:Modified vector system pCAMBIA1301m. C:Modified vector system pCAMBIA1301s. D:Schematic diagram of multi-genes interference vector system
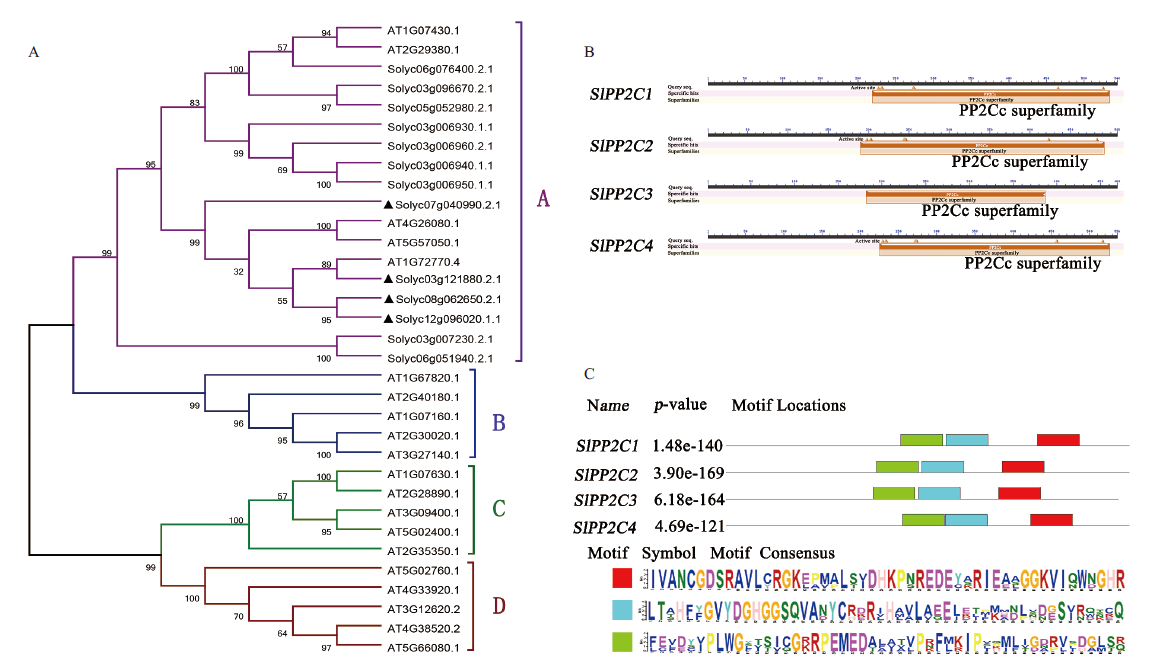
图2 四个PP2C基因的生物信息学分析 A:系统发育树;B:结构域;C:保守区域
Fig. 2 Bioinformatics analysis of four PP2C genes A:Phylogenetic tree. B:Structural domain. C:Conserved region

图3 SlPP2Cs干扰载体构建及阳性植株检测 A:SlPP2Cs多基因干扰载体系统;B:GUS染色,WT为野生型番茄,T0-1、T0-2、T0-3和T0-4为转基因番茄;C:直接PCR电泳图,M:2 kb DNA marker;1-5:显示SlPP2C1的反向片段;6-10:显示SlPP2C1的正向片段;11-15显示SlPP2C2的反向片段;16-20:显示SlPP2C2的正向片段;21-25:显示SlPP2C3的反向片段;26-30:显示SlPP2C3的正向片段;31-35:显示SlPP2C4的反向片段;36-40:显示SlPP2C4的正向片段。1-40为WT、T0-1、T0-2、T0-3和T0-4依次循环排列
Fig 3 Construction of SlPP2 Cs silencing vector and detection of positive plants A:Schematic diagram of SlPP2Cs multi-genes interference. B:GUS stain. WT:Wild-type tomato,T0-1,T0-2,T0-3 and T0-4 representing transgenic tomato. C:Direct PCR electrophoresis. M:2 kb DNA marker,1-5:showing reverse fragment of SlPP2C1,6-10:showing forward fragment of SlPP2C1;11-15:showing reverse fragment of SlPP2C2;16-20:showing forward fragment of SlPP2C2;21-25:showing reverse fragment of SlPP2C3;26-30:showing forward fragment of SlPP2C3;31-35 showing reverse fragment of SlPP2C4;36-40:showing forward fragment of SlPP2C4. 1-40 is the cyclic arrangement of WT,T0-1,T0-2,T0-3 and T0-4
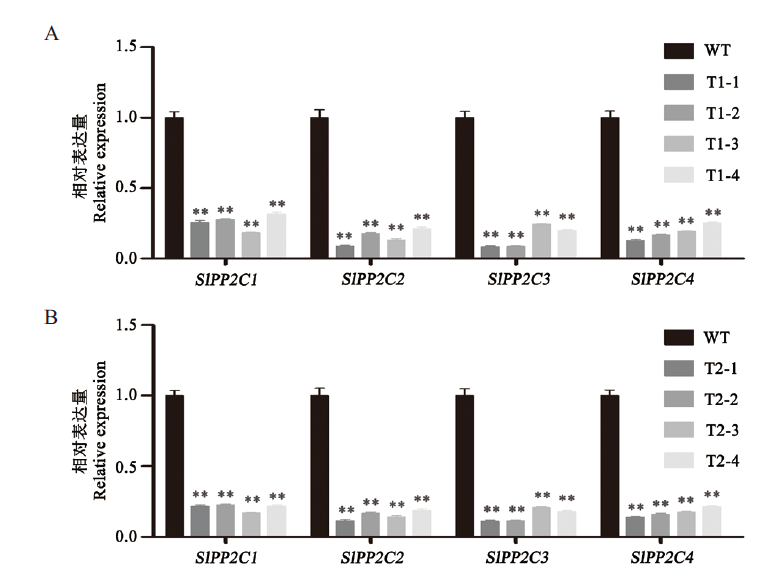
图4 四个目标基因干扰后的表达变化 A:T1代转基因植株中4个目标基因的表达变化;B:T2代转基因植株中4个目标基因的表达变化。t检验分析,*:相比于WT,P<0.05;**:相比于WT,P<0.01(WT:野生型番茄;T1-1、T1-2、T1-3、T1-4、T2-1、T2-2、T2-3和T2-4:转基因番茄),下同
Fig. 4 Expression changes of four target genes after interf-erence A:Expression changes of four target genes in T1 generation transgenic plants. B:Expression changes of four target genes in T2 generation transgenic plants. Student's t test analysis,*:Compared with WT,P<0.05. **:Compared with WT,P<0.01. WT:Wild-type,T1-1,T1-2,T1-3,T1-4,T2-1,T2-2,T2-3 and T2-4 representing transgenic tomato. The same below
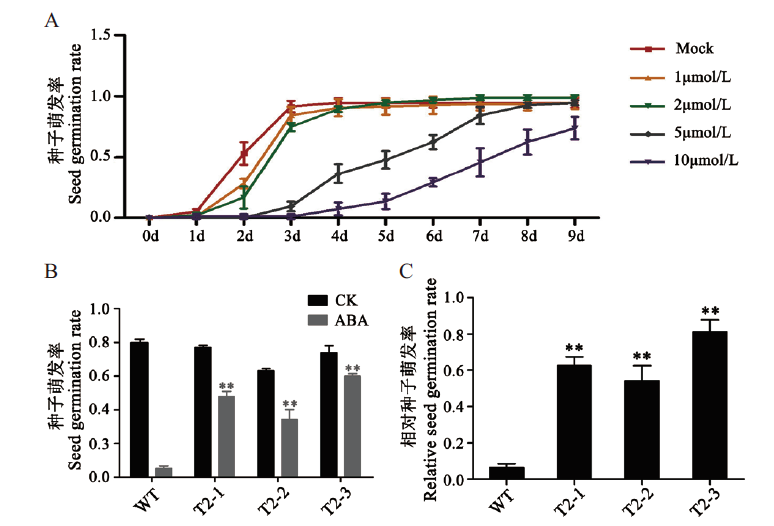
图5 转基因植株对ABA敏感性检测 A:WT对外源施加ABA的敏感性变化。横坐标代表时间,共统计9 d;B:干扰植株的种子萌发率。CK:双蒸水处理;C:干扰植株的相对种子萌发率
Fig. 5 Sensitivity test of transgenic RNAi plants to exoge-nous ABA A:Sensitivity of WT to exogenous ABA. X-axis represent time,9 d in total. B:Seed germination rate of interferred plants. CK:Double distilled water treatment. C:Relative seed germination rate of interfering plants
| [1] | Jack T. Molecular and genetic mechanisms of floral control[J]. Plant Cell, 2004, 16(Suppl):S1-S17. |
| [2] |
Ditta G, Pinyopich A, Robles P, et al. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity[J]. Curr Biol, 2004, 14(21): 1935-1940.
doi: 10.1016/j.cub.2004.10.028 pmid: 15530395 |
| [3] |
Pelaz S, Ditta GS, Baumann E, et al. B and C floral organ identity functions require SEPALLATA MADS-box genes[J]. Nature, 2000, 405(6783): 200-203.
doi: 10.1038/35012103 URL |
| [4] |
Yoshida T, Nishimura N, Kitahata N, et al. ABA-hypersensitive germination3 encodes a protein phosphatase 2C(AtPP2CA)that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs[J]. Plant Physiol, 2006, 140(1): 115-126.
pmid: 16339800 |
| [5] |
Leymarie J, Vavasseur A, Lascève G. CO2 sensing in stomata of abi1-1 and abi2-1 mutants of Arabidopsis thaliana[J]. Plant Physiol Biochem, 1998, 36(7): 539-543.
doi: 10.1016/S0981-9428(98)80180-0 URL |
| [6] |
Win J, Morgan W, Bos J, et al. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes[J]. Plant Cell, 2007, 19(8): 2349-2369.
doi: 10.1105/tpc.107.051037 pmid: 17675403 |
| [7] | 韩长志. 植物病原卵菌RxLR效应基因功能研究进展[J]. 北方园艺, 2014(5): 188-193. |
| Han CZ. Research advance on functional effect of gene plant pathogenic oomycete[J]. North Hortic, 2014(5): 188-193. | |
| [8] |
Fujita Y, Fujita M, Shinozaki K, et al. ABA-mediated transcriptional regulation in response to osmotic stress in plants[J]. J Plant Res, 2011, 124(4): 509-525.
doi: 10.1007/s10265-011-0412-3 pmid: 21416314 |
| [9] | 胡楠. 快速易用开放的植物CRISPR/Cas9系统研发[D]. 重庆: 重庆大学, 2018. |
| Hu N. Development of rapid and user-friendly open-source CRISPR/Cas9 system[D]. Chongqing: Chongqing University, 2018. | |
| [10] |
Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells[J]. Science, 2002, 296(5567): 550-553.
doi: 10.1126/science.1068999 pmid: 11910072 |
| [11] |
Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs[J]. Genes Dev, 2001, 15(2): 188-200.
doi: 10.1101/gad.862301 URL |
| [12] |
Kerschen A, Napoli CA, Jorgensen RA, et al. Effectiveness of RNA interference in transgenic plants[J]. FEBS Lett, 2004, 566(1/2/3): 223-228.
doi: 10.1016/j.febslet.2004.04.043 URL |
| [13] |
Wesley SV, Helliwell CA, Smith NA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants[J]. Plant J, 2001, 27(6): 581-590.
doi: 10.1046/j.1365-313x.2001.01105.x pmid: 11576441 |
| [14] |
Dalmay T, Hamilton A, Rudd S, et al. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus[J]. Cell, 2000, 101(5): 543-553.
doi: 10.1016/s0092-8674(00)80864-8 pmid: 10850496 |
| [15] | 徐进. 马铃薯StSUT2基因干扰载体的构建及遗传转化[D]. 兰州: 兰州理工大学, 2017. |
| Xu J. Construction of RNA interference vector of potato sucrose transporter 2 and its genetic transformation[D]. Lanzhou: Lanzhou University of Technology, 2017. | |
| [16] | 王锦达. 赤拟谷盗RNAi及dsRNA脱靶效应的研究[D]. 南京: 南京农业大学, 2015. |
| Wang JD. RNAi in Tribolium castaneum and the off target effect of dsRNA[D]. Nanjing: Nanjing Agricultural University, 2015. | |
| [17] |
Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi[J]. Nat Biotechnol, 2003, 21(6): 635-637.
doi: 10.1038/nbt831 pmid: 12754523 |
| [18] | 栾颖, 梁晋刚, 周晓莉, 等. RNAi转基因作物安全评价研究进展[J]. 生物安全学报, 2019, 28(2): 95-102. |
| Luan Y, Liang JG, Zhou XL, et al. Discussion on safety evaluation of RNAi transgenic crops[J]. J Biosaf, 2019, 28(2): 95-102. | |
| [19] |
Zhang GF, Zhao HT, Zhang CG, et al. TCP7 functions redundantly with several Class I TCPs and regulates endoreplication in Arabidopsis[J]. J Integr Plant Biol, 2019, 61(11): 1151-1170.
doi: 10.1111/jipb.12749 URL |
| [20] | 武娟. PAD4参与MPK4调控的拟南芥生长发育机制研究[D]. 北京: 中国农业大学, 2017. |
| Wu J. The regulatory mechanism of PAD4 in MPK4-regulated plant growth and development in Arabidopsis[D]. Beijing: China Agricultural University, 2017. | |
| [21] | 徐雪珍, 郑月萍, 张夏婷, 等. 拟南芥AtFAD6基因突变体的构建[J]. 江苏农业学报, 2021, 37(5): 1125-1130. |
| Xu XZ, Zheng YP, Zhang XT, et al. Construction of Arabidopsis AtFAD6 gene mutant[J]. Jiangsu J Agric Sci, 2021, 37(5): 1125-1130. | |
| [22] | 柴国华, 白泽涛, 蔡丽, 等. 油菜基因BnWRI1的克隆及RNAi对种子含油量的影响[J]. 中国农业科学, 2009, 42(5): 1512-1518. |
| Chai GH, Bai ZT, Cai L, et al. Cloning of BnWRI1 gene and the effect of RNA interference on seed oil content in oilseed rape[J]. Sci Agric Sin, 2009, 42(5): 1512-1518. | |
| [23] |
Peng Q, Hu Y, Wei R, et al. Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in Brassica napus seeds[J]. Plant Cell Rep, 2010, 29(4): 317-325.
doi: 10.1007/s00299-010-0823-y pmid: 20130882 |
| [24] |
Hüsken A, Baumert A, Strack D, et al. Reduction of sinapate ester content in transgenic oilseed rape(Brassica napus)by dsRNAi-based suppression of BnSGT1 gene expression[J]. Mol Breed, 2005, 16(2): 127-138.
doi: 10.1007/s11032-005-6825-8 URL |
| [25] |
Padmanaban S, Lin XY, Perera I, et al. Differential expression of vacuolar H+-ATPase subunit c genes in tissues active in membrane trafficking and their roles in plant growth as revealed by RNAi[J]. Plant Physiol, 2004, 134(4): 1514-1526.
doi: 10.1104/pp.103.034025 pmid: 15051861 |
| [26] |
Subramanian S, Graham MY, Yu O, et al. RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae[J]. Plant Physiol, 2005, 137(4): 1345-1353.
doi: 10.1104/pp.104.057257 pmid: 15778457 |
| [27] | 陈艳, 柴友荣, 张迪, 等. 芸薹属TT19基因家族RNA干扰载体的构建[J]. 贵州农业科学, 2011, 39(3): 1-4. |
| Chen Y, Chai YR, Zhang D, et al. Construction of RNA interference vector of TT19 gene family in Brassica[J]. Guizhou Agric Sci, 2011, 39(3): 1-4. | |
| [28] |
He F, Ni N, Zeng ZY, et al. FAMSi:a synthetic biology approach to the fast assembly of multiplex siRNAs for silencing gene expression in mammalian cells[J]. Mol Ther Nucleic Acids, 2020, 22:885-899.
doi: 10.1016/j.omtn.2020.10.007 URL |
| [29] |
Wang X, Yuan CF, Huang B, et al. Developing a versatile shotgun cloning strategy for single-vector-based multiplex expression of short interfering RNAs(siRNAs)in mammalian cells[J]. ACS Synth Biol, 2019, 8(9): 2092-2105.
doi: 10.1021/acssynbio.9b00203 pmid: 31465214 |
| [30] | 陈任, 张虹, 张芮, 等. 用于植物多基因表达载体构建的质粒系统[J]. 分子植物育种, 2018, 16(4): 1138-1146. |
| Chen R, Zhang H, Zhang R, et al. A plasmid system for construction of plant multiple gene expression vectors[J]. Mol Plant Breed, 2018, 16(4): 1138-1146. | |
| [31] | 陈立志. 椰子FATB3、LPAAT和KASI多基因载体构建和协同作用分析[D]. 海口: 海南大学, 2018. |
| Chen LZ. Multi-gene vector assembly and co-expression analysis of FatB3, LPAAT and KASI from coconut[D]. Haikou: Hainan University, 2018. |
| [1] | 刘珍银, 段郅臻, 彭婷, 王童欣, 王健. 基于三角梅的病毒诱导基因沉默体系的建立与优化[J]. 生物技术通报, 2023, 39(7): 123-130. |
| [2] | 史建磊, 宰文珊, 苏世闻, 付存念, 熊自立. 番茄青枯病抗性相关miRNA的鉴定与表达分析[J]. 生物技术通报, 2023, 39(5): 233-242. |
| [3] | 车永梅, 郭艳苹, 刘广超, 叶青, 李雅华, 赵方贵, 刘新. 菌株C8和B4的分离鉴定及其耐盐促生效果和机制[J]. 生物技术通报, 2023, 39(5): 276-285. |
| [4] | 胡明月, 杨宇, 郭仰东, 张喜春. 低温胁迫下番茄SlMYB96的功能分析[J]. 生物技术通报, 2023, 39(4): 236-245. |
| [5] | 申云鑫, 施竹凤, 周旭东, 李铭刚, 张庆, 冯路遥, 陈齐斌, 杨佩文. 三株具生防功能芽孢杆菌的分离鉴定及其生物活性研究[J]. 生物技术通报, 2023, 39(3): 267-277. |
| [6] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [7] | 陈浩婷, 张玉静, 刘洁, 代泽敏, 刘伟, 石玉, 张毅, 李天来. 低磷胁迫下番茄转录因子WRKY6功能分析[J]. 生物技术通报, 2023, 39(10): 136-147. |
| [8] | 罗皓天, 王龙, 王禹茜, 王月, 李佳祯, 杨梦珂, 张杰, 邓欣, 王红艳. 青狗尾草RNAi途径相关基因的全基因组鉴定和表达分析[J]. 生物技术通报, 2023, 39(1): 175-186. |
| [9] | 孙威, 张艳, 王聿晗, 徐僡, 徐小蓉, 鞠志刚. 马缨杜鹃Rd3GT1的克隆及对矮牵牛花色形成的影响[J]. 生物技术通报, 2022, 38(9): 198-206. |
| [10] | 陆新华, 孙德权, 张秀梅. 介孔硅纳米粒作为植物细胞转基因载体的研究[J]. 生物技术通报, 2022, 38(7): 194-204. |
| [11] | 申恒, 刘思慧, 李跃, 李敬涛, 梁文星. 一种用于PCR的番茄DNA快速粗提方法[J]. 生物技术通报, 2022, 38(6): 74-80. |
| [12] | 马馨馨, 许洋, 赵欢欢, 火兆燕, 王树彬, 钟凤林. 番茄4CL基因家族鉴定和氮素处理下的表达分析[J]. 生物技术通报, 2022, 38(4): 163-173. |
| [13] | 张鸿雁, 林国莉, 李如莲, 纪晓琦. 番茄果腐病拮抗菌的筛选及对番茄的防腐保鲜作用[J]. 生物技术通报, 2022, 38(3): 69-78. |
| [14] | 杨佳慧, 孙玉萍, 陆雅宁, 刘欢, 卢存福, 陈玉珍. 拟南芥AtTERT对大肠杆菌非生物胁迫抗性的影响[J]. 生物技术通报, 2022, 38(2): 1-9. |
| [15] | 党瑗, 李维, 苗向, 修宇, 林善枝. 山杏油体蛋白基因PsOLE4克隆及其调控油脂累积功能分析[J]. 生物技术通报, 2022, 38(11): 151-161. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||