








生物技术通报 ›› 2023, Vol. 39 ›› Issue (2): 96-106.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1171
苗淑楠( ), 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽(
), 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽( ), 李润植(
), 李润植( )
)
收稿日期:2022-09-20
出版日期:2023-02-26
发布日期:2023-03-07
作者简介:苗淑楠,女,硕士研究生,研究方向:作物遗传育种;E-mail: 基金资助:
MIAO Shu-nan( ), GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li(
), GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li( ), LI Run-zhi(
), LI Run-zhi( )
)
Received:2022-09-20
Published:2023-02-26
Online:2023-03-07
摘要:
解析大豆GmPDAT1在油脂合成和非生物胁迫应答中的功能,为大豆油脂改良和抗逆性分子育种提供新的科学参考。应用组学工具鉴定GmPDAT1,运用实时荧光定量PCR分析GmPDAT1在大豆不同组织和3种非生物胁迫的表达模式。使用酵母(Saccharomyces cerevisiae)TAG缺陷型突变体H1246检测GmPDAT1酶活性。结果表明,全基因组鉴定获得6个大豆GmPDAT1基因家族成员(GmPDAT1-A-GmPDAT1-F),除GmPDAT1-F具有8个外显子,其余5个GmPDAT1基因含6个外显子。GmPDAT1启动子区存在多个逆境胁迫响应顺式元件。GmPDAT1编码的蛋白序列长度介于582-668 aa,等电点(pI)为5.91-8.59。GmPDAT1蛋白均具有PLN02517超家族蛋白结构域,为膜结合蛋白,二级结构主要元件为α-螺旋和无规则卷曲。6个GmPDAT1蛋白聚类分为3个亚组,分别与花生AhPDAT1、小桐子JcPDAT1和蓖麻RcPDAT1-2亲缘关系较近。GmPDAT1基因家族成员具有组织特异性表达特性,其中GmPDAT1-B在不同组织均表达,且在发育种子表达量最高。推测GmPDAT1-B可能参与大豆种子TAG合成。酵母(Saccharomyces cerevisiae)TAG缺陷型突变体H1246进行功能互补测试证明,GmPDAT1-B具有催化TAG合成的酶活性。在低温、干旱和盐胁迫处理下,GmPDAT1基因家族成员呈现了不同的表达模式,预示它们可差异化参与大豆不同胁迫应答。尤其是GmPDAT1-B可介导3种不同逆境胁迫的响应。GmPDAT1-B可能具有促进大豆油脂合成和胁迫抗性的双重功能。
苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106.
MIAO Shu-nan, GAO Yu, LI Xin-ru, CAI Gui-ping, ZHANG Fei, XUE Jin-ai, JI Chun-li, LI Run-zhi. Functional Analysis of Soybean GmPDAT1 Genes in the Oil Biosynthesis and Response to Abiotic Stresses[J]. Biotechnology Bulletin, 2023, 39(2): 96-106.
| 引物名称Primer name | 引物序列 Sequence(5'-3') |
|---|---|
| GmActin-F | AGACCTTCAATGTGCCAGCCA |
| GmActin-R | CACGACCAGCAAGATCCAACC |
| qGmPDAT1-A-F | TGGCACCTTTGGGGAATTGT |
| qGmPDAT1-A-R | TCCTCATACCCAATGCGAGC |
| qGmPDAT1-B-F | CCCCCAAGATGATGAAGCGT |
| qGmPDAT1-B-R | AGGGATCCCAACCCCATACA |
| qGmPDAT1-C-F | GCCGGTTCTAAGTTCAGGCT |
| qGmPDAT1-C-R | CGTGAGCACCACTTTGTGTG |
| qGmPDAT1-D-F | ACGGAGAAAAGGGTCGGAAC |
| qGmPDAT1-D-R | CAAAGTGCAAATGCACCCCA |
| qGmPDAT1-E-F | CTGCCATGTTGCTTGGCTTT |
| qGmPDAT1-E-R | TGAAAAGGCCCTCTGCACAA |
| qGmPDAT1-F-F | TCTTGAGCGAGTTATGCGGG |
| qGmPDAT1-F-R | ATCAGTCCACACAGCCTCAC |
| GmPDAT1-B-CDs-F | ATGTCGTTTATACGACGCAGAA |
| GmPDAT1-B-CDs-R | CTAGAGCTTCAAATTGATCTTTTCA |
| GmPDAT1-B-pYES2-F | GGGGTACCATGTCGTTTATACGACGCAGAA |
| GmPDAT1-B-pYES2-R | CGGAATTCCTAGAGCTTCAAATTGATCTTTTCA |
表1 引物信息
Table 1 Primer information
| 引物名称Primer name | 引物序列 Sequence(5'-3') |
|---|---|
| GmActin-F | AGACCTTCAATGTGCCAGCCA |
| GmActin-R | CACGACCAGCAAGATCCAACC |
| qGmPDAT1-A-F | TGGCACCTTTGGGGAATTGT |
| qGmPDAT1-A-R | TCCTCATACCCAATGCGAGC |
| qGmPDAT1-B-F | CCCCCAAGATGATGAAGCGT |
| qGmPDAT1-B-R | AGGGATCCCAACCCCATACA |
| qGmPDAT1-C-F | GCCGGTTCTAAGTTCAGGCT |
| qGmPDAT1-C-R | CGTGAGCACCACTTTGTGTG |
| qGmPDAT1-D-F | ACGGAGAAAAGGGTCGGAAC |
| qGmPDAT1-D-R | CAAAGTGCAAATGCACCCCA |
| qGmPDAT1-E-F | CTGCCATGTTGCTTGGCTTT |
| qGmPDAT1-E-R | TGAAAAGGCCCTCTGCACAA |
| qGmPDAT1-F-F | TCTTGAGCGAGTTATGCGGG |
| qGmPDAT1-F-R | ATCAGTCCACACAGCCTCAC |
| GmPDAT1-B-CDs-F | ATGTCGTTTATACGACGCAGAA |
| GmPDAT1-B-CDs-R | CTAGAGCTTCAAATTGATCTTTTCA |
| GmPDAT1-B-pYES2-F | GGGGTACCATGTCGTTTATACGACGCAGAA |
| GmPDAT1-B-pYES2-R | CGGAATTCCTAGAGCTTCAAATTGATCTTTTCA |
| 蛋白 Protein | 氨基酸数目 Number of amino acids | 分子质量 Molecular weight/kD | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 亲水系数 GRAVY | 跨膜结构 Transmembrane structure |
|---|---|---|---|---|---|---|
| GmPDAT1-A | 668 | 74.833 | 8.59 | 36.12 | -0.270 | 1 |
| GmPDAT1-B | 668 | 74.776 | 8.59 | 36.87 | -0.256 | 1 |
| GmPDAT1-C | 676 | 75.571 | 6.28 | 41.96 | -0.360 | 1 |
| GmPDAT1-D | 668 | 74.557 | 6.28 | 42.50 | -0.335 | 1 |
| GmPDAT1-E | 625 | 70.376 | 6.45 | 42.51 | -0.173 | 1 |
| GmPDAT1-F | 582 | 65.783 | 5.91 | 44.95 | -0.112 | 1 |
| AtPDAT1 | 671 | 74.156 | 6.50 | 35.95 | -0.301 | 1 |
表2 大豆 Gm PDAT1蛋白质理化性质
Table 2 Physicochemical properties of GmPDAT1 proteins in G. max
| 蛋白 Protein | 氨基酸数目 Number of amino acids | 分子质量 Molecular weight/kD | 理论等电点 Theoretical pI | 不稳定系数 Instability index | 亲水系数 GRAVY | 跨膜结构 Transmembrane structure |
|---|---|---|---|---|---|---|
| GmPDAT1-A | 668 | 74.833 | 8.59 | 36.12 | -0.270 | 1 |
| GmPDAT1-B | 668 | 74.776 | 8.59 | 36.87 | -0.256 | 1 |
| GmPDAT1-C | 676 | 75.571 | 6.28 | 41.96 | -0.360 | 1 |
| GmPDAT1-D | 668 | 74.557 | 6.28 | 42.50 | -0.335 | 1 |
| GmPDAT1-E | 625 | 70.376 | 6.45 | 42.51 | -0.173 | 1 |
| GmPDAT1-F | 582 | 65.783 | 5.91 | 44.95 | -0.112 | 1 |
| AtPDAT1 | 671 | 74.156 | 6.50 | 35.95 | -0.301 | 1 |

图3 大豆GmPDAT1蛋白氨基酸序列比对 Ⅰ:盖子区域;Ⅱ:盐桥区域;Ⅲ,Ⅳ和Ⅴ:催化三联体结构。图中保守区域用红框标出
Fig. 3 Amino acid sequence alignment of GmPDAT1 proteins in G. max Ⅰ: Lid domain. Ⅱ: Salt bridge. Ⅲ, Ⅳ and Ⅴ: Catalytic triad. Conservative regions in the figure are marked with red boxes
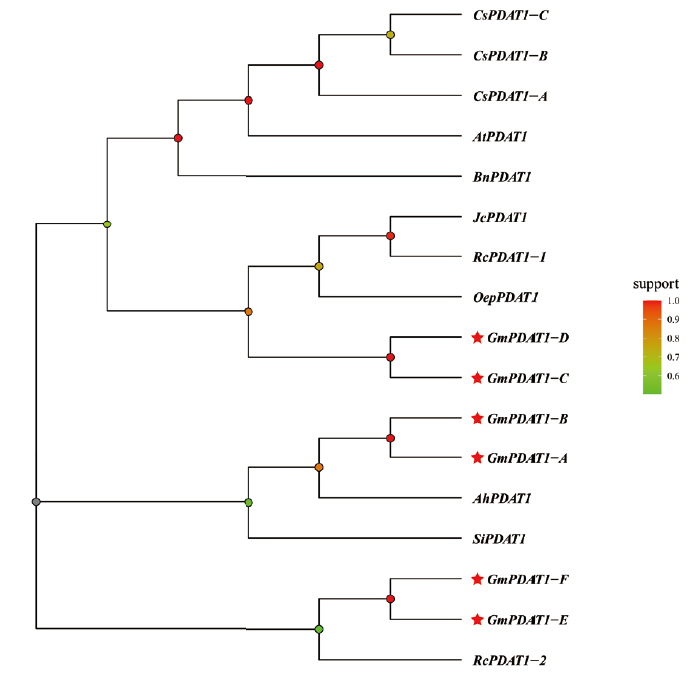
图4 大豆GmPDAT1与其他物种PDAT1的系统进化树 Gm:大豆;At:拟南芥;Rc:蓖麻;Ah:花生;Si:芝麻;Bn:甘蓝型油菜;Cs:亚麻荠;Oep:油橄榄;Jc:小桐子。颜色由红向绿代表亲缘关系由近到远
Fig. 4 Phylogenetic tree of soybean GmPDAT1s and PDA-T1s from other plant species Gm: Glycine max. At: Arabidopsis thaliana. Rc: Ricinus communis. Ah: Arachis hypogaea. Si: Sesamum indicum. Bn: Brassica napus. Cs: Camelina sativa. Oep: Olea europaea. Jc: Jatropha curcas. The color from red to green indicates the kinship from near to far

图5 大豆GmPDAT1启动子的顺式作用元件 A:顺式作用元件的数量;B:顺式作用元件的分布
Fig. 5 Cis-acting element identified in the promoters of soybean GmPDAT1 genes A: The number of cis-acting elements. B: Distribution of cis-acting elements

图7 GmPDAT1-B的酵母互补功能验证 A:转基因酵母总油脂含量(INVSc1:野生型酵母;EV:H1246转pYES2空载体;GmPDAT1-B:H1246 转pYES2-GmPDAT1-B);B:转基因酵母油脂的薄层层析(1:TLC标样;2:H1246;3:转 pYES2空载的H1246;4:野生型酵母NVSc1;5:pYES2-GmPDAT1-B转化的H1246)
Fig. 7 Complementary function assay of GmPDAT1-B gene using yeast mutant H1246 A: Total oil content of transgenic yeast(INVSc1: wild-type yeast. EV: H1246 transformed with pYES2 empty vector. GmPDAT1-B: H1246 transformed with pYES2-GmPDAT1-B). B: Thin layer chromatography of transgenic yeast oil(1: TLC standard. 2: H1246. 3: H1246 transformed with pYES2 empty vector. 4: Wild-type yeast NVSc1. 5: H1246 transformed with pYES2-GmPDAT1-B)
| [1] | 周新安. 我国大豆生产与科研现状及其发展对策[J]. 作物杂志, 2007(6): 1-4. |
| Zhou XN. The current situation of soybean production and scientific research in China and its development countermeasures[J]. Crops, 2007(6): 1-4. | |
| [2] |
Li RZ, Hatanaka T, Yu KS, et al. Soybean oil biosynthesis: role of diacylglycerol acyltransferases[J]. Funct Integr Genomics, 2013, 13(1): 99-113.
doi: 10.1007/s10142-012-0306-z URL |
| [3] |
Slocombe SP, Cornah J, Pinfield-Wells H, et al. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways[J]. Plant Biotechnol J, 2009, 7(7): 694-703.
doi: 10.1111/j.1467-7652.2009.00435.x pmid: 19702756 |
| [4] |
Kennedy EP. Biosynthesis of complex lipids[J]. Fed Proc, 1961, 20: 934-940.
pmid: 14455159 |
| [5] |
Dahlqvist A, Stahl U, Lenman M, et al. Phospholipid: diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants[J]. Proc Natl Acad Sci USA, 2000, 97(12): 6487-6492.
doi: 10.1073/pnas.120067297 pmid: 10829075 |
| [6] |
Ståhl U, Carlsson AS, Lenman M, et al. Cloning and functional characterization of a phospholipid: diacylglycerol acyltransferase from Arabidopsis[J]. Plant Physiol, 2004, 135(3): 1324-1335.
doi: 10.1104/pp.104.044354 pmid: 15247387 |
| [7] |
Fan JL, Yan CS, Zhang XB, et al. Dual role for phospholipid: diacylglycerol acyltransferase: enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves[J]. Plant Cell, 2013, 25(9): 3506-3518.
doi: 10.1105/tpc.113.117358 URL |
| [8] | 赵广, 宋志波, 刘美兰, 等. 油茶CoPDAT基因的克隆与表达分析[J]. 植物生理学报, 2017, 53(9): 1619-1628. |
| Zhao G, Song ZB, Liu ML, et al. Cloning and expression analysis of a phospholipid: diacylglycerol acyltransferase(PDAT)gene in Camellia oleifera[J]. Plant Physiol J, 2017, 53(9): 1619-1628. | |
| [9] |
Pan X, Siloto RMP, Wickramarathna AD, et al. Identification of a pair of phospholipid: diacylglycerol acyltransferases from developing flax(Linum usitatissimum L.)seed catalyzing the selective production of trilinolenin[J]. J Biol Chem, 2013, 288(33): 24173-24188.
doi: 10.1074/jbc.M113.475699 URL |
| [10] | 穆姣, 赵翠珠, 王莉莉, 等. 白菜型油菜PDAT1基因的克隆及生物信息学分析[J]. 西北农业学报, 2015, 24(12): 78-84. |
| Mu J, Zhao CZ, Wang LL, et al. Cloning and bioinformatics analysis of PDAT1 in Brassica rapa[J]. Acta Agric Boreali Occidentalis Sin, 2015, 24(12): 78-84. | |
| [11] |
谭太龙, 冯韬, 罗海燕, 等. 甘蓝型油菜磷脂二酰甘油酰基转移酶(BnPDAT1)表达特性研究[J]. 华北农学报, 2019, 34(1): 12-18.
doi: 10.7668/hbnxb.201751114 |
| Tan TL, Feng T, Luo HY, et al. Studies on the expression of phosphodiacyl glycerol acyltransferase(BnPDAT1)in Brassica napus[J]. Acta Agric Boreali Sin, 2019, 34(1): 12-18. | |
| [12] | 徐赫, 潘丽娟, 陈娜, 等. 磷脂二酰甘油酰基转移酶(PDAT)基因的克隆与表达分析[J]. 花生学报, 2018, 47(4): 33-40, 54. |
| Xu H, Pan LJ, Chen N, et al. Cloning and expression analysis of two phospholipids: diacylglycerol acyltransferase genes in peanut[J]. J Peanut Sci, 2018, 47(4): 33-40, 54. | |
| [13] | 张程, 董帅飞, 朱艺, 等. 向日葵PDAT基因家族鉴定及其对油脂积累和非生物胁迫的响应[J]. 植物生理学报, 2022, 58(5): 844-856. |
| Zhang C, Dong SF, Zhu Y, et al. Identification of sunflower PDAT gene family and their roles in TAG accumulation and abiotic stress responses[J]. Plant Physiol J, 2022, 58(5): 844-856. | |
| [14] | 张飞. 大豆GmPDAT1-B和GmDGAT3-2基因的克隆及功能分析[D]. 太谷: 山西农业大学, 2019. |
| Zhang F. Cloning and functional characterization of GmPDAT1-B and GmDGAT3-2 genes in Glycine max[D]. Taigu: Shanxi Agricultural University, 2019. | |
| [15] |
Hernández ML, Moretti S, Sicardo MD, et al. Distinct physiological roles of three phospholipid: diacylglycerol acyltransferase genes in olive fruit with respect to oil accumulation and the response to abiotic stress[J]. Front Plant Sci, 2021, 12: 751959.
doi: 10.3389/fpls.2021.751959 URL |
| [16] | 邓晓东, 蔡佳佳, 费小雯. 莱茵衣藻磷脂二脂酰甘油酰基转移酶3在三酰甘油合成中的功能研究[J]. 水生生物学报, 2014(4): 745-750. |
| Deng XD, Cai JJ, Fei XW. The role of phospholipid: diacylglycerol acyltransferase in biosynthesis of triacylglycerol by Chlamydomonas reinhardtii[J]. Acta Hydrobiol Sin, 2014(4): 745-750. | |
| [17] |
Zhang M, Fan JL, Taylor DC, et al. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development[J]. Plant Cell, 2009, 21(12): 3885-3901.
doi: 10.1105/tpc.109.071795 URL |
| [18] |
Kim HU, Lee KR, Go YS, et al. Endoplasmic Reticulum-located PDAT1-2 from Castor bean enhances hydroxy fatty acid accumulation in transgenic plants[J]. Plant Cell Physiol, 2011, 52(6): 983-993.
doi: 10.1093/pcp/pcr051 URL |
| [19] | Yuan LX, Mao X, Zhao K, et al. Characterisation of phospholipid: diacylglycerol acyltransferases(PDATs)from Camelina sativa and their roles in stress responses[J]. Biol Open, 2017, 6: 1024-1034. |
| [20] |
Mueller SP, Unger M, Guender L, et al. Phospholipid: diacylglycerol acyltransferase-mediated triacylglyerol synthesis augments basal thermotolerance[J]. Plant Physiol, 2017, 175(1): 486-497.
doi: 10.1104/pp.17.00861 URL |
| [21] |
Demski K, Łosiewska A, Jasieniecka-Gazarkiewicz K, et al. Phospholipid: diacylglycerol acyltransferase1 overexpression delays senescence and enhances post-heat and cold exposure fitness[J]. Front Plant Sci, 2020, 11: 611897.
doi: 10.3389/fpls.2020.611897 URL |
| [22] | 周雅莉, 黄旭升, 郝月茹, 等. 紫苏溶血磷脂酸酰基转移酶基因的克隆与功能分析[J]. 生物工程学报, 2022, 38(8): 3014-3028. |
| Zhou YL, Huang XS, Hao YR, et al. Cloning and functional characterization of a lysophosphatidic acid acyltransferase gene from Perilla frutescens[J]. Chin J Biotechnol, 2022, 38(8): 3014-3028. | |
| [23] |
Sandager L, Gustavsson MH, Ståhl U, et al. Storage lipid synthesis is non-essential in yeast[J]. J Biol Chem, 2002, 277(8): 6478-6482.
doi: 10.1074/jbc.M109109200 pmid: 11741946 |
| [24] |
Gasulla F, Vom Dorp K, Dombrink I, et al. The role of lipid metabolism in the acquisition of desiccation tolerance in Craterostigma plantagineum: a comparative approach[J]. Plant J, 2013, 75(5): 726-741.
doi: 10.1111/tpj.12241 URL |
| [25] |
Liu XY, Ouyang LL, Zhou ZG. Phospholipid: diacylglycerol acyltransferase contributes to the conversion of membrane lipids into triacylglycerol in Myrmecia incisa during the nitrogen starvation stress[J]. Sci Rep, 2016, 6((18)): 26610.
doi: 10.1038/srep26610 URL |
| [26] |
Guihéneuf F, Leu S, Zarka A, et al. Cloning and molecular characterization of a novel acyl-CoA: diacylglycerol acyltransferase 1-like gene(PtDGAT1)from the diatom Phaeodactylum tricornutum[J]. FEBS J, 2011, 278(19): 3651-3666.
doi: 10.1111/j.1742-4658.2011.08284.x pmid: 21812932 |
| [27] |
Gong YM, Zhang JP, Guo XJ, et al. Identification and characterization of PtDGAT2B, an acyltransferase of the DGAT2 acyl-coenzyme A: diacylglycerol acyltransferase family in the diatom Phaeodacty-lum tricornutum[J]. FEBS Lett, 2013, 587(5): 481-487.
doi: 10.1016/j.febslet.2013.01.015 URL |
| [28] |
Kong YF, Chen SB, Yang Y, et al. ABA-insensitive(ABI)4 and ABI5 synergistically regulate DGAT1 expression in Arabidopsis seedlings under stress[J]. FEBS Lett, 2013, 587(18): 3076-3082.
doi: 10.1016/j.febslet.2013.07.045 URL |
| [29] |
Farag KM, Palta JP. Use of lysophosphatidylethanolamine, a natural lipid, to retard tomato leaf and fruit senescence[J]. Physiol Plant, 1993, 87(4): 515-521.
doi: 10.1111/j.1399-3054.1993.tb02501.x URL |
| [1] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [2] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [3] | 李文辰, 刘鑫, 康越, 李伟, 齐泽铮, 于璐, 王芳. TRV病毒诱导大豆基因沉默体系优化及应用[J]. 生物技术通报, 2023, 39(7): 143-150. |
| [4] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [5] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [6] | 李苑虹, 郭昱昊, 曹燕, 祝振洲, 王飞飞. 外源植物激素调控微藻生长及目标产物积累研究进展[J]. 生物技术通报, 2023, 39(6): 61-72. |
| [7] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [8] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [9] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [10] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [11] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [12] | 侯筱媛, 车郑郑, 李姮静, 杜崇玉, 胥倩, 王群青. 大豆膜系统cDNA文库的构建及大豆疫霉效应子PsAvr3a互作蛋白的筛选[J]. 生物技术通报, 2023, 39(4): 268-276. |
| [13] | 杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142. |
| [14] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [15] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||