








生物技术通报 ›› 2023, Vol. 39 ›› Issue (4): 157-165.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0832
薛皦( ), 朱庆锋, 冯彦钊, 陈沛, 刘文华, 张爱霞, 刘勤坚, 张琪, 于洋(
), 朱庆锋, 冯彦钊, 陈沛, 刘文华, 张爱霞, 刘勤坚, 张琪, 于洋( )
)
收稿日期:2022-07-05
出版日期:2023-04-26
发布日期:2023-05-16
通讯作者:
于洋,男,博士,副研究员,研究方向:作物遗传资源发掘与创新利用;E-mail: yuyang@gdaas.cn作者简介:薛皦,女,博士,助理研究员,研究方向:植物抗病分子生物学;E-mail: xuejiao@gdaas.cn
基金资助:
XUE Jiao ZHU Qing-feng FENG Yan-zhao CHEN Pei LIU Wen-hua ZHANG Ai-xia LIU Qin-jian ZHANG Qi YU Yang( )
)
Received:2022-07-05
Published:2023-04-26
Online:2023-05-16
摘要:
上游开放阅读框(upstream open reading frame, uORF)是一类能够精确控制蛋白质翻译的mRNA元件,位于mRNA的5'端前导区,主要通过抑制翻译起始来调节下游主体开放阅读框(main open reading frame, mORF)的翻译。目前对植物uORF的预测和鉴定主要集中于生物信息学和翻译组学鉴定技术。植物uORF广泛参与调节生长发育、营养代谢、抗病免疫等多个生命活动过程。本文对植物uORF的分类、功能机制、预测和鉴定方法、植物规避uORF的方式以及植物uORF的工程应用等进行综述归纳,旨在更系统和深入地理解植物uORF的功能与机制,并为uORF应用于作物分子育种工作提供参考。
薛皦, 朱庆锋, 冯彦钊, 陈沛, 刘文华, 张爱霞, 刘勤坚, 张琪, 于洋. 植物基因上游开放阅读框的研究进展[J]. 生物技术通报, 2023, 39(4): 157-165.
XUE Jiao ZHU Qing-feng FENG Yan-zhao CHEN Pei LIU Wen-hua ZHANG Ai-xia LIU Qin-jian ZHANG Qi YU Yang. Advances in Upstream Open Reading Frame in Plant Genes[J]. Biotechnology Bulletin, 2023, 39(4): 157-165.
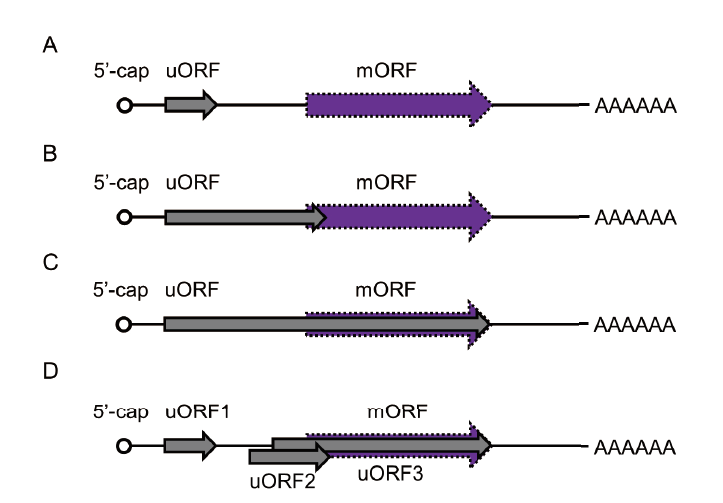
图1 植物uORF分类 A:uORF独立于mORF; B:uORF终止于mORF读码框内; C:uORF和mORF终止位置相同; D:混合型uORF
Fig. 1 Upstream open reading frame(uORF)classifica-tions in plant A: The location of the stop codon of the uORF is independent of the mORF. B: The stop codon of the uORF overlaps with the mORF. C: The stop codon of the uORF is the same as that of the stop codon of the mORF. D: Multiple uORFs
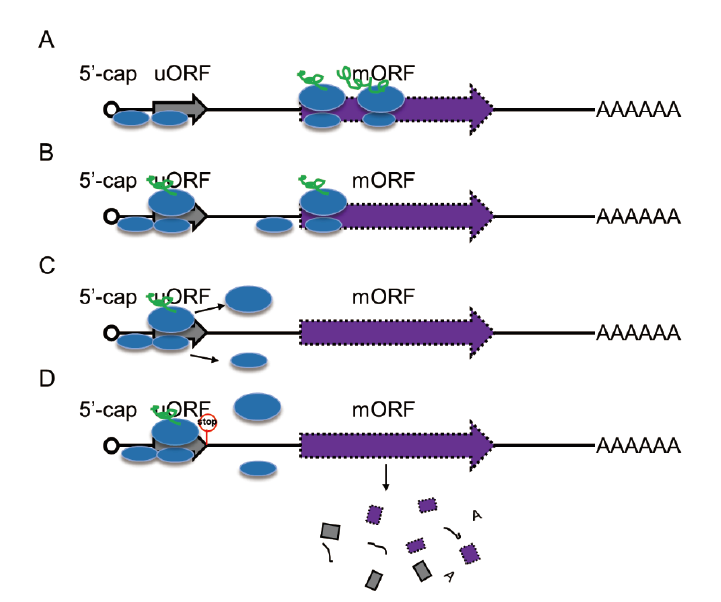
图2 植物uORF对mORF的调控方式 A:核糖体渗漏扫描; B:核糖体在mORF处重起始翻译; C:核糖体解离; D:NMD
Fig. 2 Regulatory mechanisms of uORF to mORF A: Leaky scanning by 40S ribosome subnit. B: Translational reinitiation at mORF. C: Ribosome dissociates from the mRNA; D: Trigger nonsense-mediated mRNA decay(NMD)
| 方法比较 Comparison of methods | 多聚核糖体分析技术 Polysome profiling | 核糖体-新生肽链复合物测序 RNC-Seq | 核糖体图谱分析技术 Ribo-seq | 核糖体亲和纯化技术 RAP |
|---|---|---|---|---|
| 分离原理 | 蔗糖密度梯度离心 | 单一浓度蔗糖溶液离心 | RNA酶降解未被核糖体覆盖的mRNA后经蔗糖梯度离心 | 亲和标签免疫沉淀 |
| 分离对象 | 多聚核糖体复合物 | 核糖体-新生肽链复合物 | 被核糖体覆盖保护的RNA小片段 | 核糖体大亚基及其结合的mRNA |
| 优点 | 可对核糖体数分组进行收集分析 | RNA回收容易,得率高 | 准确度高,通量大 | 可组织特异性检测 |
| 检测方法 | 高通量测序或芯片技术 | 高通量测序或芯片技术 | 高通量测序 | 高通量测序或芯片技术 |
| 检测的mRNA长度 | 全长 | 全长 | 被核糖体覆盖的约30 nt | 全长 |
| 缺点 | 仪器贵重、操作繁琐、RNA回收难度大、纯度低 | 仪器贵重、无法分析结合不同数量核糖体的 mRNA | 操作复杂、酶切条件难控制 | 需构建转基因植株 |
表1 预测uORF的实验方法比较
Table 1 Comparison of the methods for predicting uORFs
| 方法比较 Comparison of methods | 多聚核糖体分析技术 Polysome profiling | 核糖体-新生肽链复合物测序 RNC-Seq | 核糖体图谱分析技术 Ribo-seq | 核糖体亲和纯化技术 RAP |
|---|---|---|---|---|
| 分离原理 | 蔗糖密度梯度离心 | 单一浓度蔗糖溶液离心 | RNA酶降解未被核糖体覆盖的mRNA后经蔗糖梯度离心 | 亲和标签免疫沉淀 |
| 分离对象 | 多聚核糖体复合物 | 核糖体-新生肽链复合物 | 被核糖体覆盖保护的RNA小片段 | 核糖体大亚基及其结合的mRNA |
| 优点 | 可对核糖体数分组进行收集分析 | RNA回收容易,得率高 | 准确度高,通量大 | 可组织特异性检测 |
| 检测方法 | 高通量测序或芯片技术 | 高通量测序或芯片技术 | 高通量测序 | 高通量测序或芯片技术 |
| 检测的mRNA长度 | 全长 | 全长 | 被核糖体覆盖的约30 nt | 全长 |
| 缺点 | 仪器贵重、操作繁琐、RNA回收难度大、纯度低 | 仪器贵重、无法分析结合不同数量核糖体的 mRNA | 操作复杂、酶切条件难控制 | 需构建转基因植株 |
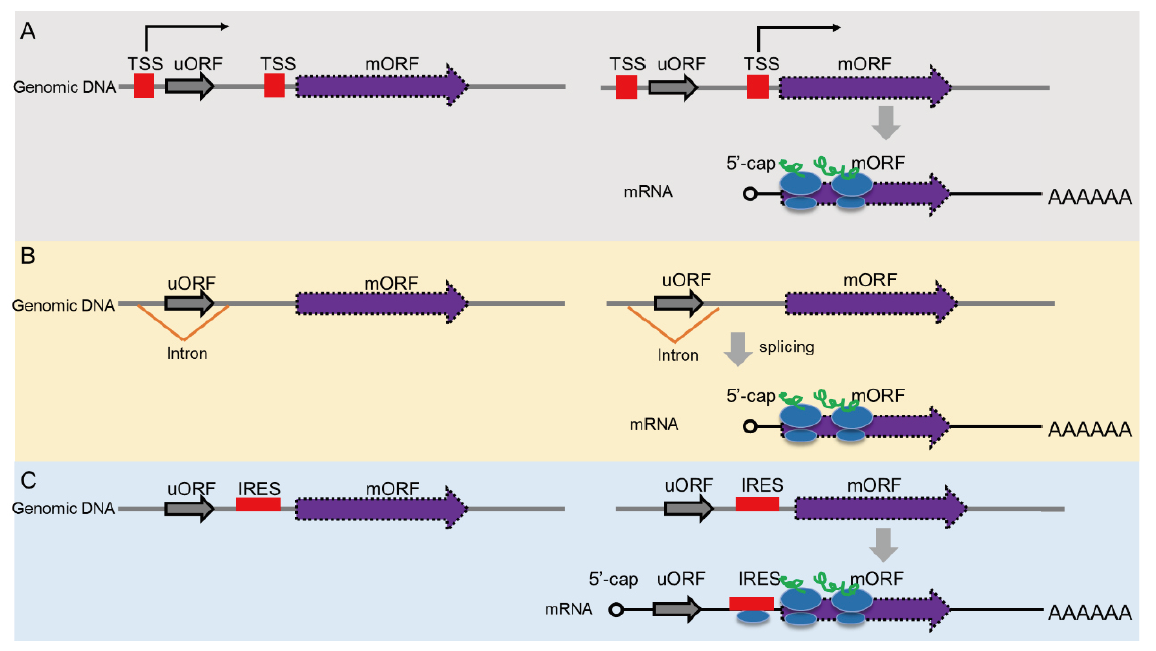
图3 植物规避uORF调节的方式 A:选择可变转录起始位点; B:可变剪切排除或修剪uORF; C:不依赖于5'帽子结构的翻译起始
Fig. 3 Mechanisms for evading uORF-mediated regulation A: Alternative transcription start site(TSS)selection. B: Processing uORF by alternative splicing. C: Cap-independent translational initiation
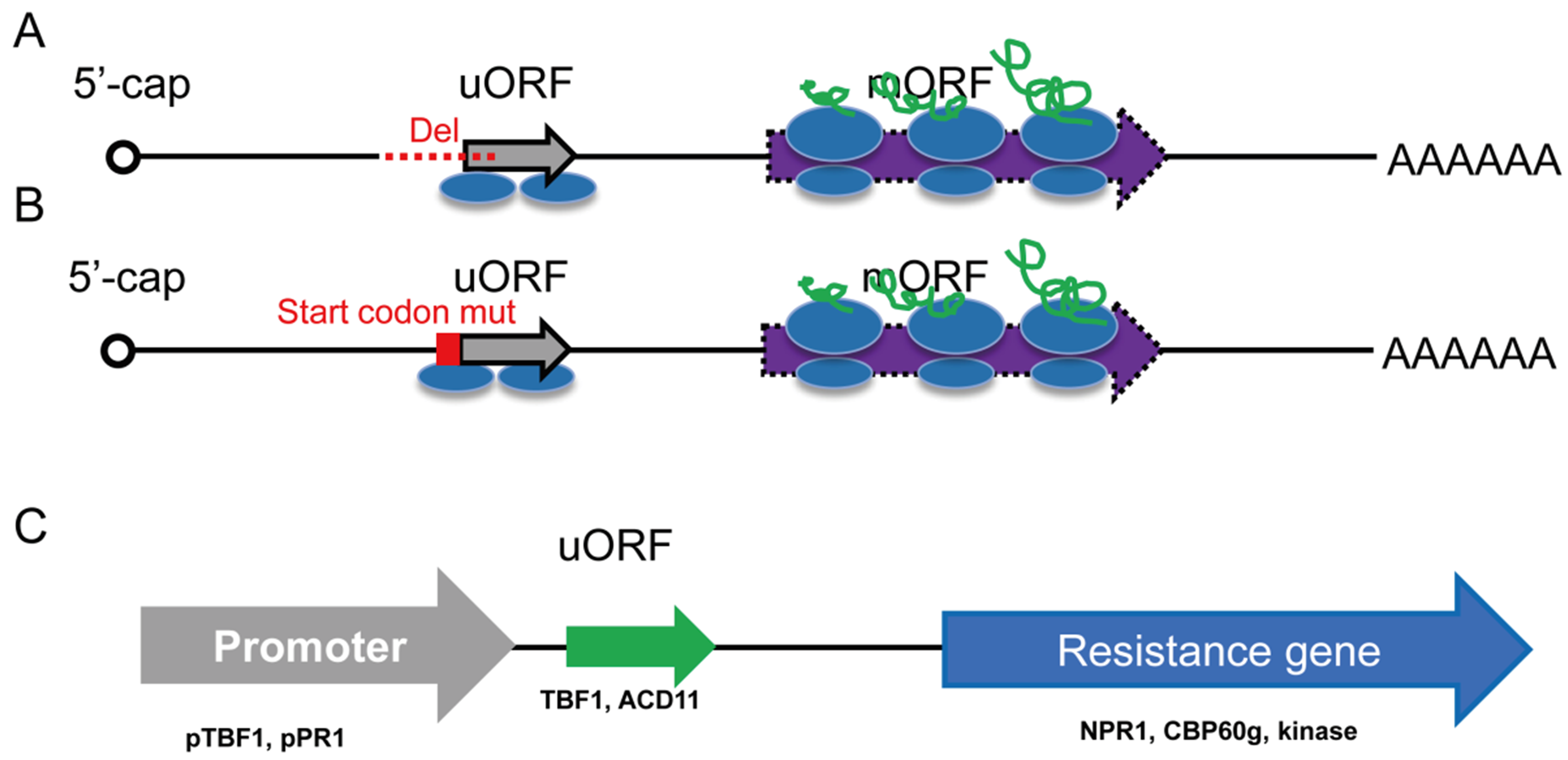
图4 应用uORF操控基因翻译 A:删除uORF上游及翻译起始序列; B:编辑突变uORF的翻译起始位点; C:基因工程手段利用uORF序列
Fig. 4 Manipulation of gene translation by editing uORFs A: Involved the deletion of uORF upstream sequence and initiation codon. B: Involved the mutation of uORF initiation codon. C: Utilized uORF sequence by genetic engineering
| [1] |
von Arnim AG, Jia QD, Vaughn JN. Regulation of plant translation by upstream open reading frames[J]. Plant Sci, 2014, 214: 1-12.
doi: 10.1016/j.plantsci.2013.09.006 pmid: 24268158 |
| [2] | Hsu PY, Calviello L, Wu HYL, et al. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis[J]. Proc Natl Acad Sci USA, 2016, 113(45): E7126-E7135. |
| [3] |
van der Horst S, Snel B, Hanson J, et al. Novel pipeline identifies new upstream ORFs and non-AUG initiating main ORFs with conserved amino acid sequences in the 5' leader of mRNAs in Arabidopsis thaliana[J]. RNA, 2019, 25(3): 292-304.
doi: 10.1261/rna.067983.118 URL |
| [4] | Hsu PY, Benfey PN. Small but mighty: functional peptides encoded by small ORFs in plants[J]. Proteomics, 2018, 18(10): e1700038. |
| [5] |
Ohtani M, Wachter A. NMD-based gene regulation-A strategy for fitness enhancement in plants?[J]. Plant Cell Physiol, 2019, 60(9): 1953-1960.
doi: 10.1093/pcp/pcz090 pmid: 31111919 |
| [6] |
Kurihara Y, Matsui A, Hanada K, et al. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis[J]. Proc Natl Acad Sci USA, 2009, 106(7): 2453-2458.
doi: 10.1073/pnas.0808902106 pmid: 19181858 |
| [7] | 贾晓波, 胡剑. 无义介导的mRNA降解[J]. 中国生物化学与分子生物学报, 2012, 28(2): 115-120. |
| Jia XB, Hu J. Nonsense-meditated mRNA decay[J]. Chin J Biochem Mol Biol, 2012, 28(2): 115-120. | |
| [8] |
Hellens RP, Brown CM, Chisnall MAW, et al. The emerging world of small ORFs[J]. Trends Plant Sci, 2016, 21(4): 317-328.
doi: S1360-1385(15)00285-X pmid: 26684391 |
| [9] |
Zhang H, Wang YR, Lu J. Function and evolution of upstream ORFs in eukaryotes[J]. Trends Biochem Sci, 2019, 44(9): 782-794.
doi: S0968-0004(19)30055-6 pmid: 31003826 |
| [10] |
Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression[J]. Wiley Interdiscip Rev RNA, 2014, 5(6): 765-778.
doi: 10.1002/wrna.1245 URL |
| [11] | 金勇丰, 边腾飞, 周萍. 高等植物基因上游可译框架(uORF)的分析[J]. 农业生物技术学报, 2004, 12(5): 493-499. |
| Jin YF, Bian TF, Zhou P. Upstream open reading frames(uORF)analysis of plant mRNAs[J]. J Agric Biotechnol, 2004, 12(5): 493-499. | |
| [12] |
Bhushan S, Meyer H, Starosta AL, et al. Structural basis for translational stalling by human Cytomegalovirus and fungal arginine attenuator peptide[J]. Mol Cell, 2010, 40(1): 138-146.
doi: 10.1016/j.molcel.2010.09.009 pmid: 20932481 |
| [13] |
Hayden CA, Jorgensen RA. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes[J]. BMC Biol, 2007, 5: 32.
doi: 10.1186/1741-7007-5-32 |
| [14] |
Tanaka M, Sotta N, Yamazumi Y, et al. The minimum open reading frame, AUG-stop, induces boron-dependent ribosome stalling and mRNA degradation[J]. Plant Cell, 2016, 28(11): 2830-2849.
doi: 10.1105/tpc.16.00481 URL |
| [15] |
Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation[J]. Nat Rev Mol Cell Biol, 2010, 11(2): 113-127.
doi: 10.1038/nrm2838 |
| [16] |
Rahmani F, Hummel M, Schuurmans J, et al. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide[J]. Plant Physiol, 2009, 150(3): 1356-1367.
doi: 10.1104/pp.109.136036 pmid: 19403731 |
| [17] |
Yamashita Y, Takamatsu S, Glasbrenner M, et al. Sucrose sensing through nascent peptide-meditated ribosome stalling at the stop Codon of Arabidopsis bZIP11 uORF2[J]. FEBS Lett, 2017, 591(9): 1266-1277.
doi: 10.1002/1873-3468.12634 pmid: 28369795 |
| [18] |
Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5'-untranslated regions of eukaryotic mRNAs[J]. Science, 2016, 352(6292): 1413-1416.
doi: 10.1126/science.aad9868 pmid: 27313038 |
| [19] |
Uchiyama-Kadokura N, Murakami K, Takemoto M, et al. Polyamine-responsive ribosomal arrest at the stop Codon of an upstream open reading frame of the AdoMetDC1 gene triggers nonsense-mediated mRNA decay in Arabidopsis thaliana[J]. Plant Cell Physiol, 2014, 55(9): 1556-1567.
doi: 10.1093/pcp/pcu086 pmid: 24929422 |
| [20] | Niu RX, Zhou YL, Zhang Y, et al. uORFlight: a vehicle toward uORF-mediated translational regulation mechanisms in eukaryotes[J]. Database(Oxford), 2020, 2020: baaa007. |
| [21] |
Chen YJ, Li DY, Fan WL, et al. PsORF: a database of small ORFs in plants[J]. Plant Biotechnol J, 2020, 18(11): 2158-2160.
doi: 10.1111/pbi.v18.11 URL |
| [22] |
Hazarika RR, de Coninck B, Yamamoto LR, et al. ARA-PEPs: a repository of putative sORF-encoded peptides in Arabidopsis thaliana[J]. BMC Bioinformatics, 2017, 18(1): 37.
doi: 10.1186/s12859-016-1458-y pmid: 28095775 |
| [23] |
Wiese A, Elzinga N, Wobbes B, et al. A conserved upstream open reading frame mediates sucrose-induced repression of translation[J]. Plant Cell, 2004, 16(7): 1717-1729.
doi: 10.1105/tpc.019349 pmid: 15208401 |
| [24] |
Tran MK, Schultz CJ, Baumann U. Conserved upstream open reading frames in higher plants[J]. BMC Genomics, 2008, 9: 361.
doi: 10.1186/1471-2164-9-361 pmid: 18667093 |
| [25] |
Zhao J, Qin B, Nikolay R, et al. Translatomics: the global view of translation[J]. Int J Mol Sci, 2019, 20(1): 212.
doi: 10.3390/ijms20010212 URL |
| [26] |
Cai CW, Wang P, Zhao CY, et al. The use of ribosome-nascent chain complex-seq to reveal the translated mRNA profile and the role of ASN1 in resistance to Verticillium wilt in cotton[J]. Genomics, 2021, 113(6): 3872-3880.
doi: 10.1016/j.ygeno.2021.09.015 URL |
| [27] |
Halbeisen RE, Scherrer T, Gerber AP. Affinity purification of ribosomes to access the translatome[J]. Methods, 2009, 48(3): 306-310.
doi: 10.1016/j.ymeth.2009.04.003 pmid: 19398006 |
| [28] |
Ingolia NT. Ribosome footprint profiling of translation throughout the genome[J]. Cell, 2016, 165(1): 22-33.
doi: S0092-8674(16)30216-1 pmid: 27015305 |
| [29] |
Calviello L, Mukherjee N, Wyler E, et al. Detecting actively translated open reading frames in ribosome profiling data[J]. Nat Methods, 2016, 13(2): 165-170.
doi: 10.1038/nmeth.3688 pmid: 26657557 |
| [30] |
Erhard F, Halenius A, Zimmermann C, et al. Improved Ribo-seq enables identification of cryptic translation events[J]. Nat Methods, 2018, 15(5): 363-366.
doi: 10.1038/nmeth.4631 pmid: 29529017 |
| [31] |
Kurihara Y, Makita Y, Kawashima M, et al. Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis[J]. Proc Natl Acad Sci USA, 2018, 115(30): 7831-7836.
doi: 10.1073/pnas.1804971115 pmid: 29915080 |
| [32] |
Tokizawa M, Kusunoki K, Koyama H, et al. Identification of Arabidopsis genic and non-genic promoters by paired-end sequencing of TSS tags[J]. Plant J, 2017, 90(3): 587-605.
doi: 10.1111/tpj.2017.90.issue-3 URL |
| [33] |
Srivastava AK, Lu YM, Zinta G, et al. UTR-dependent control of gene expression in plants[J]. Trends Plant Sci, 2018, 23(3): 248-259.
doi: S1360-1385(17)30255-8 pmid: 29223924 |
| [34] |
Pasentsis K, Paulo N, Algarra P, et al. Characterization and expression of the phytochrome gene family in the moss Ceratodon purpureus[J]. Plant J, 1998, 13(1): 51-61.
pmid: 9680964 |
| [35] |
Combier JP, de Billy F, Gamas P, et al. Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development[J]. Genes Dev, 2008, 22(11): 1549-1559.
doi: 10.1101/gad.461808 URL |
| [36] |
Lee KM, Chen CJ, Shih SR. Regulation mechanisms of viral IRES-driven translation[J]. Trends Microbiol, 2017, 25(7): 546-561.
doi: 10.1016/j.tim.2017.01.010 URL |
| [37] |
Yamamoto YY, Tsuhara Y, Gohda K, et al. Gene trapping of the Arabidopsis genome with a firefly luciferase reporter[J]. Plant J, 2003, 35(2): 273-283.
doi: 10.1046/j.1365-313X.2003.01797.x URL |
| [38] |
Kurihara Y, Okubo-Kurihara E, Matsui M. Polycistronic expression of RNA silencing suppressor protects its own mRNA from RNA silencing[J]. Plant Biotechnol, 2015, 32: 89-95.
doi: 10.5511/plantbiotechnology.15.0120b URL |
| [39] |
Tiburcio AF, Alcázar R. Potential applications of polyamines in agriculture and plant biotechnology[J]. Methods Mol Biol, 2018, 1694: 489-508.
doi: 10.1007/978-1-4939-7398-9_40 pmid: 29080190 |
| [40] |
Hanfrey C, Elliott KA, Franceschetti M, et al. A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation[J]. J Biol Chem, 2005, 280(47): 39229-39237.
doi: 10.1074/jbc.M509340200 pmid: 16176926 |
| [41] |
Guerrero-González ML, Ortega-Amaro MA, Juárez-Montiel M, et al. Arabidopsis polyamine oxidase-2 uORF is required for downstream translational regulation[J]. Plant Physiol Biochem, 2016, 108: 381-390.
doi: 10.1016/j.plaphy.2016.08.006 URL |
| [42] |
Guerrero-González ML, Rodríguez-Kessler M, Jiménez-Bremont JF. uORF, a regulatory mechanism of the Arabidopsis polyamine oxidase 2[J]. Mol Biol Rep, 2014, 41(4): 2427-2443.
doi: 10.1007/s11033-014-3098-5 pmid: 24435979 |
| [43] |
Salazar-Díaz K, Dong YH, Papdi C, et al. TOR senses and regulates spermidine metabolism during seedling establishment and growth in maize and Arabidopsis[J]. iScience, 2021, 24(11): 103260.
doi: 10.1016/j.isci.2021.103260 URL |
| [44] |
Cruz-Ramírez A, López-Bucio J, Ramírez-Pimentel G, et al. The xipotl mutant of Arabidopsis reveals a critical role for phospholipid metabolism in root system development and epidermal cell integrity[J]. Plant Cell, 2004, 16(8): 2020-2034.
pmid: 15295103 |
| [45] |
Alatorre-Cobos F, Cruz-Ramírez A, Hayden CA, et al. Translational regulation of Arabidopsis XIPOTL1 is modulated by phosphocholine levels via the phylogenetically conserved upstream open reading frame 30[J]. J Exp Bot, 2012, 63(14): 5203-5221.
doi: 10.1093/jxb/ers180 pmid: 22791820 |
| [46] |
Tanaka M, Takano J, Chiba Y, et al. Boron-dependent degradation of NIP5;1 mRNA for acclimation to excess boron conditions in Arabidopsis[J]. Plant Cell, 2011, 23(9): 3547-3559.
doi: 10.1105/tpc.111.088351 URL |
| [47] |
Yang SY, Lu WC, Ko SS, et al. Upstream open reading frame and phosphate-regulated expression of rice OsNLA1 controls phosphate transport and reproduction[J]. Plant Physiol, 2020, 182(1): 393-407.
doi: 10.1104/pp.19.01101 URL |
| [48] |
Guo ZL, Cao HR, Zhao J, et al. A natural uORF variant confers phosphorus acquisition diversity in soybean[J]. Nat Commun, 2022, 13(1): 3796.
doi: 10.1038/s41467-022-31555-2 pmid: 35778398 |
| [49] |
Thalor SK, Berberich T, Lee SS, et al. Deregulation of sucrose-controlled translation of a bZIP-type transcription factor results in sucrose accumulation in leaves[J]. PLoS One, 2012, 7(3): e33111.
doi: 10.1371/journal.pone.0033111 URL |
| [50] |
Laing WA, Martínez-Sánchez M, Wright MA, et al. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis[J]. Plant Cell, 2015, 27(3): 772-786.
doi: 10.1105/tpc.114.133777 URL |
| [51] |
Nishimura T, Wada T, Yamamoto KT, et al. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning[J]. Plant Cell, 2005, 17(11): 2940-2953.
pmid: 16227452 |
| [52] |
Ribone PA, Capella M, Arce AL, et al. A uORF represses the transcription factor AtHB1 in aerial tissues to avoid a deleterious phenotype[J]. Plant Physiol, 2017, 175(3): 1238-1253.
doi: 10.1104/pp.17.01060 pmid: 28956754 |
| [53] |
Dong J, Chen HD, Deng XW, et al. Phytochrome B induces intron retention and translational inhibition of PHYTOCHROME-INTERACTING FACTOR3[J]. Plant Physiol, 2020, 182(1): 159-166.
doi: 10.1104/pp.19.00835 pmid: 31690709 |
| [54] |
Pajerowska-Mukhtar KM, Wang W, Tada Y, et al. The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition[J]. Curr Biol, 2012, 22(2): 103-112.
doi: 10.1016/j.cub.2011.12.015 pmid: 22244999 |
| [55] |
Ai G, Fu X, Li TL, et al. Making use of plant uORFs to control transgene translation in response to pathogen attack[J]. BioDesign Research, 2022, 2022: 9820540. DOI: 10.34133/2022/9820540.
doi: 10.34133/2022/9820540 |
| [56] |
Wu HW, Fajiculay E, Wu JF, et al. Noise reduction by upstream open reading frames[J]. Nat Plants, 2022, 8(5): 474-480.
doi: 10.1038/s41477-022-01136-8 |
| [57] |
Zhang HW, Si XM, Ji X, et al. Genome editing of upstream open reading frames enables translational control in plants[J]. Nat Biotechnol, 2018, 36(9): 894-898.
doi: 10.1038/nbt.4202 pmid: 30080209 |
| [58] |
Si XM, Zhang HW, Wang YP, et al. Manipulating gene translation in plants by CRISPR-Cas9-mediated genome editing of upstream open reading frames[J]. Nat Protoc, 2020, 15(2): 338-363.
doi: 10.1038/s41596-019-0238-3 pmid: 31915386 |
| [59] |
Xing SN, Chen KL, Zhu HC, et al. Fine-tuning sugar content in strawberry[J]. Genome Biol, 2020, 21(1): 230.
doi: 10.1186/s13059-020-02146-5 pmid: 32883370 |
| [60] |
Xu GY, Yuan M, Ai CR, et al. uORF-mediated translation allows engineered plant disease resistance without fitness costs[J]. Nature, 2017, 545(7655): 491-494.
doi: 10.1038/nature22372 URL |
| [61] |
Kim JH, Castroverde CDM, Huang S, et al. Increasing the resilience of plant immunity to a warming climate[J]. Nature, 2022, 607(7918): 339-344.
doi: 10.1038/s41586-022-04902-y |
| [1] | 胡海琳, 徐黎, 李晓旭, 王晨璨, 梅曼, 丁文静, 赵媛媛. 小肽激素调控植物生长发育及逆境生理研究进展[J]. 生物技术通报, 2023, 39(7): 13-25. |
| [2] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [3] | 魏明, 王欣玉, 伍国强, 赵萌. NAD依赖型去乙酰化酶SRT在植物表观遗传调控中的作用[J]. 生物技术通报, 2023, 39(4): 59-70. |
| [4] | 桑田, 王鹏程. 植物SUMO化修饰研究进展[J]. 生物技术通报, 2023, 39(3): 1-12. |
| [5] | 孙雨桐, 刘德帅, 齐迅, 冯美, 黄栩筝, 姚文孔. 茉莉酸调控植物生长发育和胁迫的研究进展[J]. 生物技术通报, 2023, 39(11): 99-109. |
| [6] | 安昌, 陆琳, 沈梦千, 陈盛圳, 叶康卓, 秦源, 郑平. 植物bHLH基因家族研究进展及在药用植物中的应用前景[J]. 生物技术通报, 2023, 39(10): 1-16. |
| [7] | 汤茜茜, 林楚宇, 陶增. 植物组蛋白去甲基化酶研究进展[J]. 生物技术通报, 2022, 38(7): 13-22. |
| [8] | 李萍, 郭发平, 田敏, 税阳, 徐娜娜, 白大嵩, 余德金, 张杰, 胡运高, 彭友林. 甾醇在调节植物生长发育中的研究进展[J]. 生物技术通报, 2022, 38(7): 90-98. |
| [9] | 古盼, 齐学影, 李莉, 张曦, 单晓昳. AtRGS1胞吞动态调控G蛋白参与拟南芥生长发育和抗性反应[J]. 生物技术通报, 2022, 38(6): 34-42. |
| [10] | 李兵娟, 郑璐, 沈仁芳, 兰平. 拟南芥RPP1A参与幼苗生长的蛋白质组学分析[J]. 生物技术通报, 2022, 38(2): 10-20. |
| [11] | 悦曼芳, 张春, 吴忠义. 植物转录因子AP2/ERF家族蛋白结构和功能的研究进展[J]. 生物技术通报, 2022, 38(12): 11-26. |
| [12] | 马荣, 尚方正, 潘剑锋, 戎友俊, 王敏, 李金泉, 张燕军. 细胞内mRNA翻译影响因素及翻译组学的研究进展[J]. 生物技术通报, 2022, 38(12): 115-126. |
| [13] | 汤晓丽, 姜福东, 张洪霞. 植物SINA E3泛素连接酶功能的研究进展[J]. 生物技术通报, 2022, 38(10): 10-17. |
| [14] | 钱静洁, 林苏梦, 张冬平, 高勇. 光敏色素互作因子参与生长素调控的植物生长发育[J]. 生物技术通报, 2022, 38(10): 29-33. |
| [15] | 苏雨, 李宗芸, 韩永华. 植物液泡加工酶研究进展[J]. 生物技术通报, 2021, 37(6): 181-191. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||