








生物技术通报 ›› 2023, Vol. 39 ›› Issue (9): 117-125.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0090
收稿日期:2023-02-07
出版日期:2023-09-26
发布日期:2023-10-24
通讯作者:
张蕾,女,博士,副教授,研究方向:植物发育生物学;E-mail: arabilab@whu.edu.cn作者简介:王子颖,女,硕士研究生,研究方向:植物发育生物学;E-mail: 747262414@qq.com
基金资助:
WANG Zi-ying( ), LONG Chen-jie, FAN Zhao-yu, ZHANG Lei(
), LONG Chen-jie, FAN Zhao-yu, ZHANG Lei( )
)
Received:2023-02-07
Published:2023-09-26
Online:2023-10-24
摘要:
水稻是重要的粮食作物,研究水稻生长发育调控机制可为水稻品种改良奠定理论基础。钙依赖蛋白激酶(calcium dependent protein kinases, CDPKs)是植物中重要的蛋白激酶,参与植物生长发育以及对环境反应的应答。水稻中的CRK5(CDPK-related kinase 5)在蛋白序列和结构上与CDPK高度同源。为进一步研究OsCRK5在水稻干旱反应中的功能,本研究利用酵母双杂交筛库技术筛选了OsCRK5的互作蛋白。首先将OsCRK5的1-1 332 bp片段克隆至pGBKT7载体中,获得诱饵载体pGBKT7-OsCRK5,经测序无误后,转化至酵母菌株Y2H Gold中。在营养缺陷培养基中观察到重组蛋白不具有毒性作用及自激活活性,同时利用Western blot分析重组蛋白的表达。进一步利用水稻cDNA文库筛选OsCRK5的互作蛋白,共得到77个阳性克隆。功能预测结果显示,互作蛋白涉及蛋白质合成、贮存和降解过程、转录调控、植物细胞生长和分裂过程、能量代谢和细胞代谢等方面的功能。最后,从阳性克隆中选取参与植物干旱胁迫应答的两个蛋白OsWR1和OsDi19-1,利用酵母双杂交和双分子荧光互补的方法验证其与OsCRK5的相互作用。研究结果为水稻抗旱的遗传改良奠定了基础。
王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125.
WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System[J]. Biotechnology Bulletin, 2023, 39(9): 117-125.
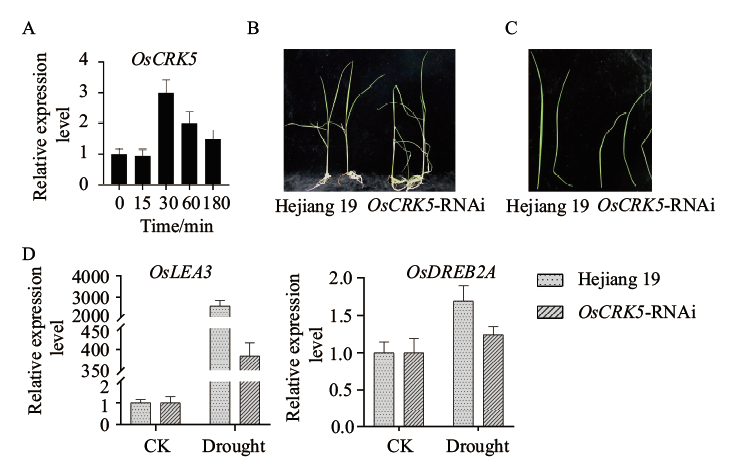
图1 OsCRK5参与水稻干旱胁迫反应 A:定量PCR检测PEG处理后水稻叶片中OsCRK5的表达情况;B:20% PEG处理20 d水稻幼苗的表型;C:20% PEG处理水稻幼苗后叶片的表型;D:定量PCR检测干旱诱导表达基因在野生型和OsCRK5-RNAi植株中的表达情况
Fig. 1 OsCRK5 participates in the response of rice to drought stress A: The expression of OsCRK5 in the leaves after PEG treatment by qPCR detection. B: Phenotype of 20 d rice seedlings treated with 20% PEG. C: Leaf phenotypes of rice seedlings treated with 20% PEG. D: Quantitative PCR to detect the expressions of drought-induced genes in wild-type and OsCRK5-RNAi plants
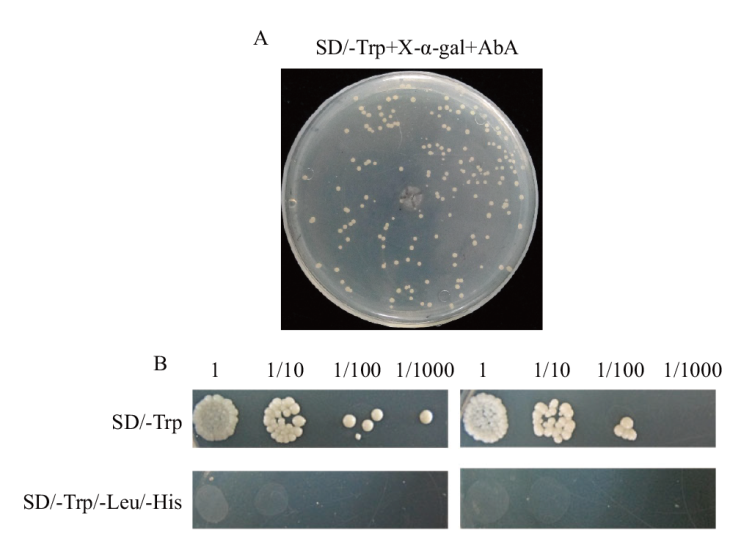
图2 pGBKT7-OsCRK51-444质粒在酵母细胞中的自激活检测 A:pGBKT7-OsCRK51-444质粒转化的酵母细胞在SD/Trp+X-α-gal培养基上的生长状况;B:pGBKT7-OsCRK51-444质粒转化的酵母细胞在单缺及三缺固体培养基上的生长状况。以pGBKT7空载质粒转化的酵母细胞生长状况为对照
Fig. 2 Detection of self-activation of plasmid pGBKT7-OsCRK51-444 in yeast cells A: Growth condition of the yeast cells transformed with pGBKT7-OsCRK51-444 plasmid on SD/Trp+X-α-gal plate. B: Growth condition of the yeast cells transformed with pGBKT7-OsCRK51-444 plasmid on single and triple deficiency plates. The growth of yeast transformed with pGBKT7 plasmid was used as control
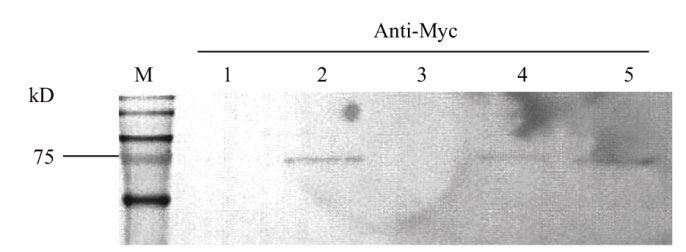
图3 不同酵母菌株中诱导蛋白表达水平的检测 1-5为不同酵母菌株中提取的总蛋白
Fig. 3 Detection of expressions of trait protein in different yeast strains 1-5 were total proteins isolated from different yeast strains
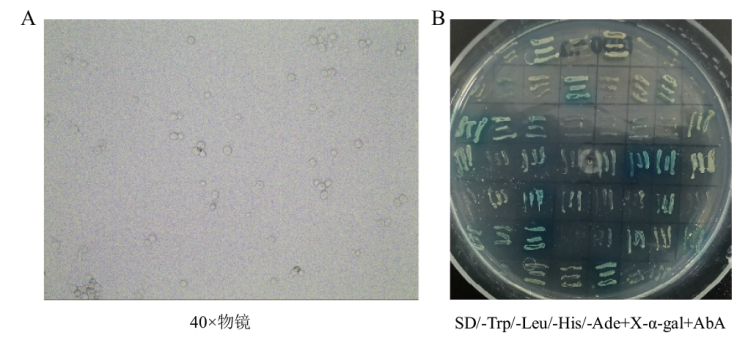
图4 筛选获得的部分阳性克隆 A:40倍物镜下观察到的酵母合子;B:SD/-Trp/-Leu/-His/-Ade+X-α-gal+AbA培养基上二次筛选得到的部分阳性菌斑
Fig. 4 Partial positive clones from screened A: Yeast zygote observed under 40× objective lens. B: Partial positive clones screened from SD/-Trp/- Leu/- His/- Ade+X- α-gal+AbA plate
| NCBI号 Accession number of NCBI | 基因名称 Gene name | 基因注释 Gene annotation | 参考文献 Reference |
|---|---|---|---|
| LOC4347054 | ZHD9 | 锌指同源结构域蛋白9 Zinc-finger homeodomain protein 9-like | [ |
| LOC107275681 | ARP5 | 肌动蛋白相关蛋白5 Actin-related protein 5 | [ |
| LOC4339608 | Di19-1 | 脱水诱导蛋白19 DEHYDRATION-INDUCED 19-like protein | [ |
| LOC4336195 | ATP23 | 线粒体内膜蛋白酶 Mitochondrial inner membrane protease | [ |
| LOC4329772 | OsFWL2 | 细胞数目调节因子2 Cell number regulator 2 | [ |
| LOC4337552 | OseIF3f | 真核翻译起始因子3 Eukaryotic translation initiation factor 3 subunit F | [ |
| LOC4329963 | OsCBP20 | 核帽结合蛋白亚基2 Nuclear cap-binding protein subunit 2-like | [ |
| LOC4349349 | OsEXPB3 | 类膨胀素B3 Expansin-B3-like | [ |
| LOC4334101 | rcs3 | 类半胱氨酸合成酶 Cysteine synthase-like | [ |
| LOC4345839 | OsMtATPd2 | 线粒体中d亚单位ATP合成酶 ATP synthase subunit d, mitochondrial | [ |
| LOC4325989 | BBTI8 | 胰蛋白酶抑制剂 Bowman-Birk type bran trypsin inhibitor | [ |
| LOC4331740 | OsCCR4b | 碳分解代谢抑制蛋白4同源物1 Carbon catabolite repressor protein 4 homolog 1 | [ |
| LOC4349455 | BSK2 | 丝氨酸/苏氨酸蛋白激酶 Serine/threonine-protein kinase | [ |
| LOC4336306 | OsHCT1 | 羟基肉桂酰转移酶 Hydroxycinnamoyltransferase 1-like | [ |
| LOC4330573 | BSH | 染色体结构重塑蛋白 Chromatin structure-remodeling complex protein | |
| LOC4340286 | HPGT3 | 羟脯氨酸O-半乳糖基转移酶 Hydroxyproline O-galactosyltransferase | [ |
| LOC9269857 | OsWRKY29 | WRKY转录因子29 Probable WRKY transcription factor 29 | [ |
| LOC4349610 | OsWRKY70 | WRKY转录因子70 Probable WRKY transcription factor 70 | [ |
| LOC4337733 | OsSAMS1 | 类S-腺苷甲硫氨酸合酶1 S-adenosylmethionine synthase 1-like | [ |
| LOC4339460 | OsNAP1;2 | 核小体组装蛋白1;2 Nucleosome assembly protein 1;2-like | |
| LOC4328653 | OsWR1 | 乙烯应答转录因子WIN1 Ethylene-responsive transcription factor WIN1 | [ |
表1 酵母文库筛选部分互作蛋白信息
Table 1 Information of part interacting proteins screened by yeast library
| NCBI号 Accession number of NCBI | 基因名称 Gene name | 基因注释 Gene annotation | 参考文献 Reference |
|---|---|---|---|
| LOC4347054 | ZHD9 | 锌指同源结构域蛋白9 Zinc-finger homeodomain protein 9-like | [ |
| LOC107275681 | ARP5 | 肌动蛋白相关蛋白5 Actin-related protein 5 | [ |
| LOC4339608 | Di19-1 | 脱水诱导蛋白19 DEHYDRATION-INDUCED 19-like protein | [ |
| LOC4336195 | ATP23 | 线粒体内膜蛋白酶 Mitochondrial inner membrane protease | [ |
| LOC4329772 | OsFWL2 | 细胞数目调节因子2 Cell number regulator 2 | [ |
| LOC4337552 | OseIF3f | 真核翻译起始因子3 Eukaryotic translation initiation factor 3 subunit F | [ |
| LOC4329963 | OsCBP20 | 核帽结合蛋白亚基2 Nuclear cap-binding protein subunit 2-like | [ |
| LOC4349349 | OsEXPB3 | 类膨胀素B3 Expansin-B3-like | [ |
| LOC4334101 | rcs3 | 类半胱氨酸合成酶 Cysteine synthase-like | [ |
| LOC4345839 | OsMtATPd2 | 线粒体中d亚单位ATP合成酶 ATP synthase subunit d, mitochondrial | [ |
| LOC4325989 | BBTI8 | 胰蛋白酶抑制剂 Bowman-Birk type bran trypsin inhibitor | [ |
| LOC4331740 | OsCCR4b | 碳分解代谢抑制蛋白4同源物1 Carbon catabolite repressor protein 4 homolog 1 | [ |
| LOC4349455 | BSK2 | 丝氨酸/苏氨酸蛋白激酶 Serine/threonine-protein kinase | [ |
| LOC4336306 | OsHCT1 | 羟基肉桂酰转移酶 Hydroxycinnamoyltransferase 1-like | [ |
| LOC4330573 | BSH | 染色体结构重塑蛋白 Chromatin structure-remodeling complex protein | |
| LOC4340286 | HPGT3 | 羟脯氨酸O-半乳糖基转移酶 Hydroxyproline O-galactosyltransferase | [ |
| LOC9269857 | OsWRKY29 | WRKY转录因子29 Probable WRKY transcription factor 29 | [ |
| LOC4349610 | OsWRKY70 | WRKY转录因子70 Probable WRKY transcription factor 70 | [ |
| LOC4337733 | OsSAMS1 | 类S-腺苷甲硫氨酸合酶1 S-adenosylmethionine synthase 1-like | [ |
| LOC4339460 | OsNAP1;2 | 核小体组装蛋白1;2 Nucleosome assembly protein 1;2-like | |
| LOC4328653 | OsWR1 | 乙烯应答转录因子WIN1 Ethylene-responsive transcription factor WIN1 | [ |
| [1] | 李琼, 姜雪. 水稻产量相关性状的遗传研究[J]. 安徽农业科学, 2022, 50(13): 10-13, 20. |
| Li Q, Jiang X. Genetic studies on rice yield related traits[J]. J Anhui Agric Sci, 2022, 50(13): 10-13, 20. | |
| [2] | 李涵茂, 陆魁东, 戴平, 等. 不同品种双季晚稻产量结构对干旱胁迫的响应[J]. 气象与环境科学, 2019, 42(2): 27-34. |
| Li HM, Lu KD, Dai P, et al. Research on yield structure of different varieties of double-cropping late rice response to drought stress[J]. Meteorol Environ Sci, 2019, 42(2): 27-34. | |
| [3] |
Asano T, Hayashi N, Kikuchi S, et al. CDPK-mediated abiotic stress signaling[J]. Plant Signal Behav, 2012, 7(7): 817-821.
doi: 10.4161/psb.20351 pmid: 22751324 |
| [4] |
Boudsocq M, Sheen J. CDPKs in immune and stress signaling[J]. Trends Plant Sci, 2013, 18(1): 30-40.
doi: 10.1016/j.tplants.2012.08.008 pmid: 22974587 |
| [5] | Cheng SH, Willmann MR, Chen HC, et al. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family[J]. Plant Physiol, 2002, 129(2): 469-485. |
| [6] |
Asano T, Tanaka N, Yang G, et al. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice[J]. Plant Cell Physiol, 2005, 46(2): 356-366.
doi: 10.1093/pcp/pci035 pmid: 15695435 |
| [7] |
Atif RM, Shahid L, Waqas M, et al. Insights on calcium-dependent protein kinases(CPKs)signaling for abiotic stress tolerance in plants[J]. Int J Mol Sci, 2019, 20(21): 5298.
doi: 10.3390/ijms20215298 URL |
| [8] |
Li AL, Zhu YF, Tan XM, et al. Evolutionary and functional study of the CDPK gene family in wheat(Triticum aestivum L.)[J]. Plant Mol Biol. 2008, 66(4): 429-443.
doi: 10.1007/s11103-007-9281-5 URL |
| [9] |
Xu WW, Huang WC. Calcium-dependent protein kinases in phytohormone signaling pathways[J]. Int J Mol Sci, 2017, 18(11): 2436.
doi: 10.3390/ijms18112436 URL |
| [10] |
Yip Delormel T, Boudsocq M. Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana[J]. New Phytol, 2019, 224(2): 585-604.
doi: 10.1111/nph.16088 pmid: 31369160 |
| [11] |
Durian G, Sedaghatmehr M, Matallana-Ramirez LP, et al. Calcium-dependent protein kinase CPK1 controls cell death by in vivo phosphorylation of senescence master regulator ORE1[J]. Plant Cell, 2020, 32(5): 1610-1625.
doi: 10.1105/tpc.19.00810 URL |
| [12] | Marques J, Matiolli CC, Abreu IA. Visualization of a curated Oryza sativa L. CDPKs protein-protein interaction network(CDPK-OsPPIN)[J]. MicroPubl Biol, 2022. DOI: 10.17912/micropub.biology.000513. |
| [13] |
Zhang HF, Wei CH, Yang XZ, et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its related kinase gene families in melon(Cucumis melo L.)[J]. PLoS One, 2017, 12(4): e0176352.
doi: 10.1371/journal.pone.0176352 URL |
| [14] |
Zuo R, Hu R, Chai G, et al. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar(Populus trichocarpa)[J]. Mol Biol Rep, 2013, 40(3): 2645-2662.
doi: 10.1007/s11033-012-2351-z URL |
| [15] | Cai H, Cheng J, Yan Y, et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its closely related kinase genes in Capsicum annuum[J]. Front Plant Sci, 2015, 6: 737. |
| [16] |
Wu P, Wang WL, Duan WK, et al. Comprehensive analysis of the CDPK-SnRK superfamily genes in Chinese cabbage and its evolutionary implications in plants[J]. Front Plant Sci, 2017, 8: 162.
doi: 10.3389/fpls.2017.00162 pmid: 28239387 |
| [17] |
Wang JP, Xu YP, Munyampundu JP, et al. Calcium-dependent protein kinase(CDPK)and CDPK-related kinase(CRK)gene families in tomato: genome-wide identification and functional analyses in disease resistance[J]. Mol Genet Genomics, 2016, 291(2): 661-676.
doi: 10.1007/s00438-015-1137-0 URL |
| [18] |
Tao XC, Lu YT. Loss of AtCRK1 gene function in Arabidopsis thaliana decreases tolerance to salt[J]. J Plant Biol, 2013, 56(5): 306-314.
doi: 10.1007/s12374-012-0352-z URL |
| [19] |
Baba AI, Rigó G, Ayaydin F, et al. Functional analysis of the Arabidopsis thaliana CDPK-related kinase family: AtCRK1 regulates responses to continuous light[J]. Int J Mol Sci, 2018, 19(5): 1282.
doi: 10.3390/ijms19051282 URL |
| [20] |
Nemoto K, Ramadan A, Arimura GI, et al. Tyrosine phosphorylation of the GARU E3 ubiquitin ligase promotes gibberellin signalling by preventing GID1 degradation[J]. Nat Commun, 2017, 8(1): 1004.
doi: 10.1038/s41467-017-01005-5 pmid: 29042542 |
| [21] |
Li RJ, Hua W, Lu YT. Arabidopsis cytosolic glutamine synthetase AtGLN1;1 is a potential substrate of AtCRK3 involved in leaf senescence[J]. Biochem Biophys Res Commun, 2006, 342(1): 119-126.
doi: 10.1016/j.bbrc.2006.01.100 URL |
| [22] |
Rigó G, Ayaydin F, Tietz O, et al. Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis[J]. Plant Cell, 2013, 25(5): 1592-1608.
doi: 10.1105/tpc.113.110452 URL |
| [23] |
Yadav A, Garg T, Singh H, et al. Tissue-specific expression pattern of calcium-dependent protein kinases-related kinases(CRKs)in rice[J]. Plant Signal Behav, 2020, 15(11): 1809846.
doi: 10.1080/15592324.2020.1809846 URL |
| [24] |
Fields S, Song OK. A novel genetic system to detect protein-protein interactions[J]. Nature, 1989, 340(6230): 245-246.
doi: 10.1038/340245a0 |
| [25] |
Chien CT, Bartel PL, Sternglanz R, et al. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest[J]. Proc Natl Acad Sci USA, 1991, 88(21): 9578-9582.
pmid: 1946372 |
| [26] |
de Folter S, Immink RGH. Yeast protein-protein interaction assays and screens[J]. Methods Mol Biol, 2011, 754: 145-165.
doi: 10.1007/978-1-61779-154-3_8 pmid: 21720951 |
| [27] |
Bollier N, Gonzalez N, Chevalier C, et al. Zinc finger-homeodomain and mini zinc finger proteins are key players in plant growth and responses to environmental stresses[J]. J Exp Bot, 2022, 73(14): 4662-4673.
doi: 10.1093/jxb/erac194 URL |
| [28] |
Yao W, Beckwith SL, Zheng TN, et al. Assembly of the Arp5(actin-related protein)subunit involved in distinct INO80 chromatin remodeling activities[J]. J Biol Chem, 2015, 290(42): 25700-25709.
doi: 10.1074/jbc.M115.674887 URL |
| [29] |
Rodriguez Milla MA, Uno Y, Chang IF, et al. A novel yeast two-hybrid approach to identify CDPK substrates: characterization of the interaction between AtCPK11 and AtDi19, a nuclear zinc finger protein 1[J]. FEBS Lett, 2006, 580(3): 904-911.
pmid: 16438971 |
| [30] |
Yang GY, Zhao T, Lu S, et al. T1121G point mutation in the mitochondrial gene COX1 suppresses a null mutation in ATP23 required for the assembly of yeast mitochondrial ATP synthase[J]. Int J Mol Sci, 2022, 23(4): 2327.
doi: 10.3390/ijms23042327 URL |
| [31] |
Thibivilliers S, Farmer A, Libault M. Biological and cellular functions of the microdomain-associated FWL/CNR protein family in plants[J]. Plants, 2020, 9(3): 377.
doi: 10.3390/plants9030377 URL |
| [32] | Li Q, Deng ZY, Gong CY, et al. The rice eukaryotic translation initiation factor 3 subunit f(OseIF3f)is involved in microgametogenesis[J]. Front Plant Sci, 2016, 7: 532. |
| [33] |
Calero G, Wilson KF, Ly T, et al. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex[J]. Nat Struct Biol, 2002, 9(12): 912-917.
doi: 10.1038/nsb874 pmid: 12434151 |
| [34] |
Schmidt R, Schippers JHM, Mieulet D, et al. MULTIPASS, a rice R2R3-type MYB transcription factor, regulates adaptive growth by integrating multiple hormonal pathways[J]. Plant J, 2013, 76(2): 258-273.
doi: 10.1111/tpj.2013.76.issue-2 URL |
| [35] |
Nakamura T, Yamaguchi Y, Sano H. Four rice genes encoding cysteine synthase: isolation and differential responses to sulfur, nitrogen and light[J]. Gene, 1999, 229(1/2): 155-161.
doi: 10.1016/S0378-1119(99)00019-0 URL |
| [36] |
Moriguchi K, Suzuki T, Ito Y, et al. Functional isolation of novel nuclear proteins showing a variety of subnuclear localizations[J]. Plant Cell, 2005, 17(2): 389-403.
pmid: 15659629 |
| [37] |
Shan SH, Yin RP, Shi JY, et al. Bowman-birk major type trypsin inhibitor derived from foxtail millet bran attenuate atherosclerosis via remodeling gut microbiota in ApoE-/-mice[J]. J Agric Food Chem, 2022, 70(2): 507-519.
doi: 10.1021/acs.jafc.1c05747 URL |
| [38] |
Chou WL, Chung YL, Fang JC, et al. Novel interaction between CCR4 and CAF1 in rice CCR4-NOT deadenylase complex[J]. Plant Mol Biol, 2017, 93(1): 79-96.
doi: 10.1007/s11103-016-0548-6 URL |
| [39] |
Tang WQ, Kim TW, Oses-Prieto JA, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis[J]. Science, 2008, 321(5888): 557-560.
doi: 10.1126/science.1156973 URL |
| [40] |
Jantasuriyarat C, Gowda M, Haller K, et al. Large-scale identification of expressed sequence tags involved in rice and rice blast fungus interaction[J]. Plant Physiol, 2005, 138(1): 105-115.
pmid: 15888683 |
| [41] |
Ogawa-Ohnishi M, Matsubayashi Y. Identification of three potent hydroxyproline O-galactosyltransferases in Arabidopsis[J]. Plant J, 2015, 81(5): 736-746.
doi: 10.1111/tpj.2015.81.issue-5 URL |
| [42] |
Zhou CL, Lin QB, Lan J, et al. WRKY transcription factor OsWRKY29 represses seed dormancy in rice by weakening abscisic acid response[J]. Front Plant Sci, 2020, 11: 691.
doi: 10.3389/fpls.2020.00691 URL |
| [43] |
Tang JQ, Mei EY, He ML, et al. Functions of OsWRKY24, OsWRKY70 and OsWRKY53 in regulating grain size in rice[J]. Planta, 2022, 255(4): 1-8.
doi: 10.1007/s00425-021-03785-z |
| [44] | Zhao SS, Hong W, Wu JG, et al. A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection[J]. eLife, 2017, 6: 27529. |
| [45] |
Wang YH, Wan LY, Zhang LX, et al. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice[J]. Plant Mol Biol, 2012, 78(3): 275-288.
doi: 10.1007/s11103-011-9861-2 pmid: 22130861 |
| [46] |
Rodriguez Milla MA, Townsend J, Chang IF, et al. The Arabidopsis AtDi19 gene family encodes a novel type of Cys2/His2 zinc-finger protein implicated in ABA-independent dehydration, high-salinity stress and light signaling pathways[J]. Plant Mol Biol, 2006, 61(1): 13-30.
doi: 10.1007/s11103-005-5798-7 URL |
| [47] |
Su L, Yang J, Li DD, et al. Dynamic genome-wide association analysis and identification of candidate genes involved in anaerobic germination tolerance in rice[J]. Rice, 2021, 14(1): 1.
doi: 10.1186/s12284-020-00444-x pmid: 33409869 |
| [48] |
Liu WX, Zhang FC, Zhang WZ, et al. Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress[J]. Mol Plant, 2013, 6(5): 1487-1502.
doi: 10.1093/mp/sst031 URL |
| [49] |
Qin LX, Li Y, Li DD, et al. Arabidopsis drought-induced protein Di19-3 participates in plant response to drought and high salinity stresses[J]. Plant Mol Biol, 2014, 86(6): 609-625.
doi: 10.1007/s11103-014-0251-4 pmid: 25218132 |
| [50] |
Qin LX, Nie XY, Hu R, et al. Phosphorylation of serine residue modulates cotton Di19-1 and Di19-2 activities for responding to high salinity stress and abscisic acid signaling[J]. Sci Rep, 2016, 6: 20371.
doi: 10.1038/srep20371 |
| [51] | Guo YL, Tan YQ, Qu MH, et al. OsWR2 recruits OsHDA704 to regulate the deacetylation of H4K8ac in the promoter of OsABI5 in response to drought stress[J]. J Integr Plant Biol, 2023. DOI: 10.1111/jipb.13481. |
| [52] |
Luban J, Goff SP. The yeast two-hybrid system for studying protein-protein interactions[J]. Curr Opin Biotechnol, 1995, 6(1): 59-64.
doi: 10.1016/0958-1669(95)80010-7 URL |
| [1] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [2] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [3] | 刘雯锦, 马瑞, 刘升燕, 杨江伟, 张宁, 司怀军. 马铃薯StCIPK11的克隆及响应干旱胁迫分析[J]. 生物技术通报, 2023, 39(9): 147-155. |
| [4] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [5] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| [6] | 吴元明, 林佳怡, 柳雨汐, 李丹婷, 张宗琼, 郑晓明, 逄洪波. 基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J]. 生物技术通报, 2023, 39(8): 173-184. |
| [7] | 姚莎莎, 王晶晶, 王俊杰, 梁卫红. 植物激素信号通路调控水稻粒型的分子机制[J]. 生物技术通报, 2023, 39(8): 80-90. |
| [8] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [9] | 丁凯鑫, 王立春, 田国奎, 王海艳, 李凤云, 潘阳, 庞泽, 单莹. 烯效唑缓解植物干旱损伤的研究进展[J]. 生物技术通报, 2023, 39(6): 1-11. |
| [10] | 王春语, 李政君, 王平, 张丽霞. 高粱表皮蜡质缺失突变体sb1抗旱生理生化分析[J]. 生物技术通报, 2023, 39(5): 160-167. |
| [11] | 任沛东, 彭健玲, 刘圣航, 姚姿婷, 朱桂宁, 陆光涛, 李瑞芳. 沙福芽孢杆菌GX-H6的分离鉴定及对水稻细菌性条斑病的防病效果[J]. 生物技术通报, 2023, 39(5): 243-253. |
| [12] | 李怡君, 吴晨晨, 李睿, 王喆, 何山文, 韦善君, 张晓霞. 水稻内生细菌新资源分离培养方案探究[J]. 生物技术通报, 2023, 39(4): 201-211. |
| [13] | 侯筱媛, 车郑郑, 李姮静, 杜崇玉, 胥倩, 王群青. 大豆膜系统cDNA文库的构建及大豆疫霉效应子PsAvr3a互作蛋白的筛选[J]. 生物技术通报, 2023, 39(4): 268-276. |
| [14] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [15] | 王琪, 胡哲, 富薇, 李光哲, 郝林. 伯克霍尔德氏菌GD17对黄瓜幼苗耐干旱的调节[J]. 生物技术通报, 2023, 39(3): 163-175. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||