








生物技术通报 ›› 2024, Vol. 40 ›› Issue (5): 167-178.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1092
侯雅琼1( ), 郎红珊1, 闻蒙蒙1, 谷易云2, 朱润洁2, 汤晓丽1(
), 郎红珊1, 闻蒙蒙1, 谷易云2, 朱润洁2, 汤晓丽1( )
)
收稿日期:2023-11-20
出版日期:2024-05-26
发布日期:2024-06-13
通讯作者:
汤晓丽,女,博士,副教授,研究方向:果树生理和逆境分子生物学;E-mail: tangxiaoli.1@163.com作者简介:侯雅琼,女,硕士研究生,研究方向:园艺植物分子模块育种;E-mail: houyaqiong2023@163.com
基金资助:
HOU Ya-qiong1( ), LANG Hong-shan1, WEN Meng-meng1, GU Yi-yun2, ZHU Run-jie2, TANG Xiao-li1(
), LANG Hong-shan1, WEN Meng-meng1, GU Yi-yun2, ZHU Run-jie2, TANG Xiao-li1( )
)
Received:2023-11-20
Published:2024-05-26
Online:2024-06-13
摘要:
【目的】鉴定分析了猕猴桃的小热休克蛋白(HSP20s/sHSPs)基因家族,为猕猴桃AcHSP20基因的生物学功能研究和猕猴桃的抗性育种奠定基础。【方法】基于猕猴桃全基因组数据,利用生物信息学方法对猕猴桃AcHSP20基因家族进行鉴定,并分析了家族成员的理化性质、系统进化、染色体定位、基因结构、亚细胞定位及启动子。同时利用荧光定量PCR技术分析了猕猴桃AcHSP20基因在非生物胁迫和激素处理后的表达变化情况。【结果】鉴定获得34个AcHSP20基因家族成员。其编码蛋白的氨基酸数目为85-371,分子量介于9.93-40.18 kD,等电点介于4.4-9.3,AcHSP20s多定位于细胞质和叶绿体。34个基因分布在17条染色体上,多数没有或只有一个内含子。基因的启动子含有植物激素和非生物胁迫响应元件。所有测定的AcHSP20基因在猕猴桃的根茎叶中均有表达且在高温及其他多种非生物胁迫和植物激素处理下差异表达显著。【结论】AcHSP20基因家族成员在猕猴桃的高温、ABA、MeJA、NaCl等逆境胁迫响应中发挥重要作用。
侯雅琼, 郎红珊, 闻蒙蒙, 谷易云, 朱润洁, 汤晓丽. 猕猴桃AcHSP20基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 167-178.
HOU Ya-qiong, LANG Hong-shan, WEN Meng-meng, GU Yi-yun, ZHU Run-jie, TANG Xiao-li. Identification and Expression Analysis of AcHSP20 Gene Family in Kiwifruit[J]. Biotechnology Bulletin, 2024, 40(5): 167-178.
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| AcHSP20-2 | GATCTATCGCACCGTGTC | AACCTTTATGTCGTCCTTG |
| AcHSP20-5 | CTTCCAGGAGGTGTATCGT | CGTCTTCTTTGGCATTGTA |
| AcHSP20-9 | TCTCACCTTTGGGTTTATTG | CAGGGAACGTCATGGAA |
| AcHSP20-12 | CGGGAGGAGGAGAAAGAA | CAAACAGCAGACACGGC |
| AcHSP20-19 | AGAAGAATGGAGGTGATGAT | GAGATGGAGCACAAGAGTTT |
| AcActin | TGCATGAGCGATCAAGTTTCAAG | TGTCCCATGTCTGGTTGATGACT |
表1 AcHSP20s表达分析的引物
Table 1 Primers used in the expression analysis of AcHSP20s genes
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| AcHSP20-2 | GATCTATCGCACCGTGTC | AACCTTTATGTCGTCCTTG |
| AcHSP20-5 | CTTCCAGGAGGTGTATCGT | CGTCTTCTTTGGCATTGTA |
| AcHSP20-9 | TCTCACCTTTGGGTTTATTG | CAGGGAACGTCATGGAA |
| AcHSP20-12 | CGGGAGGAGGAGAAAGAA | CAAACAGCAGACACGGC |
| AcHSP20-19 | AGAAGAATGGAGGTGATGAT | GAGATGGAGCACAAGAGTTT |
| AcActin | TGCATGAGCGATCAAGTTTCAAG | TGTCCCATGTCTGGTTGATGACT |
| 基因 Gene | 基因组登录号 Gene ID | 等电点 pI | 分子量 Molecular weight/kD | 氨基酸数Number of amino acids | 染色体定位及基因方向Chromosome localization and gene orientation | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|
| AcHSP20-1 | Ach00g218421.2 | 6.57 | 10.16 | 88 | Chr00: 54153215.. 54154904(-) | 细胞质Cytoplasmic |
| AcHSP20-2 | Ach00g239181 | 7.86 | 16.14 | 143 | Chr00: 50739806.. 50740562(-) | 过氧化物酶体Peroxisomal |
| AcHSP20-3 | Ach00g329621 | 5.59 | 23.28 | 208 | Chr00: 90841963.. 90843153(-) | 叶绿体Chloroplast |
| AcHSP20-4 | Ach00g479741.2 | 10.32 | 9.93 | 86 | Chr00: 135211617.. 135211877(+) | 细胞质Cytoplasmic |
| AcHSP20-5 | Ach00g479761.2 | 9.14 | 40.18 | 371 | Chr00: 145805825.. 145808080(-) | 高尔基体Golgi apparatus |
| AcHSP20-6 | Ach02g036831 | 7.8 | 16.56 | 152 | Chr02: 4423490.. 4423948(-) | 细胞质Cytoplasmic |
| AcHSP20-7 | Ach02g046741.2 | 6.21 | 25.55 | 225 | Chr02: 516872.. 518080(-) | 叶绿体Chloroplast |
| AcHSP20-8 | Ach02g223291 | 8.86 | 17.61 | 153 | Chr02: 4090770.. 4091231(-) | 细胞质Cytoplasmic |
| AcHSP20-9 | Ach03g045661 | 7.63 | 25.69 | 232 | Chr03: 6797048.. 6798489(-) | 叶绿体Chloroplast |
| AcHSP20-10 | Ach03g451751.2 | 9.15 | 11.12 | 96 | Chr03: 1045510.. 1045800(-) | 叶绿体Chloroplast |
| AcHSP20-11 | Ach04g161211.2 | 6.45 | 24.77 | 215 | Chr04: 6179073.. 6180486(-) | 细胞质Cytoplasmic |
| AcHSP20-12 | Ach06g418911.2 | 6.32 | 16.60 | 152 | Chr06: 2925504.. 2925962(-) | 细胞质Cytoplasmic |
| AcHSP20-13 | Ach10g027441.2 | 5.4 | 20.33 | 181 | Chr10: 4989172.. 4989717(-) | 细胞质Cytoplasmic |
| AcHSP20-14 | Ach10g027451 | 7.77 | 25.72 | 228 | Chr10: 4973842.. 4974528(-) | 细胞质Cytoplasmic |
| AcHSP20-15 | Ach10g027461.2 | 6.34 | 24.95 | 221 | Chr10: 4984511.. 4985176(+) | 叶绿体Chloroplast |
| AcHSP20-16 | Ach10g143161 | 9.3 | 25.78 | 228 | Chr10: 11814664.. 11815350(+) | 细胞质Cytoplasmic |
| AcHSP20-17 | Ach11g263431.2 | 5.05 | 21.72 | 196 | Chr11: 8776984.. 8777574(-) | 细胞质Cytoplasmic |
| AcHSP20-18 | Ach11g425231.2 | 4.43 | 10.59 | 90 | Chr11: 12263315.. 12263587(-) | 细胞质Cytoplasmic |
| AcHSP20-19 | Ach12g058351.2 | 8.69 | 17.21 | 152 | Chr12: 6955029.. 6956940(+) | 细胞核Nucleus |
| AcHSP20-20 | Ach13g308811 | 5.94 | 33.07 | 297 | Chr13: 1116598.. 1117580(+) | 叶绿体Chloroplast |
| AcHSP20-21 | Ach13g350901 | 5.68 | 21.90 | 195 | Chr13: 14397259.. 14398212(+) | 叶绿体Chloroplast |
| AcHSP20-22 | Ach13g383281 | 6.31 | 16.61 | 152 | Chr13: 11985660.. 11986118(-) | 细胞质Cytoplasmic |
| AcHSP20-23 | Ach13g472651.2 | 5.94 | 33.07 | 297 | Chr13: 1497079.. 1498061(-) | 叶绿体Chloroplast |
| AcHSP20-24 | Ach14g205801.2 | 6.87 | 24.06 | 216 | Chr14: 9422059.. 9423375(+) | 叶绿体Chloroplast |
| AcHSP20-25 | Ach14g205811.2 | 8.86 | 36.76 | 329 | Chr14: 9424474.. 9428179(+) | 叶绿体Chloroplast |
| AcHSP20-26 | Ach15g376651 | 5.67 | 22.90 | 202 | Chr15: 4758959.. 4759567(-) | 细胞质Cytoplasmic |
| AcHSP20-27 | Ach16g309411.2 | 7.64 | 26.14 | 231 | Chr16: 8508810.. 8510240(-) | 叶绿体Chloroplast |
| AcHSP20-28 | Ach16g331111 | 8.83 | 17.36 | 153 | Chr16: 10507526.. 10508507(+) | 细胞质Cytoplasmic |
| AcHSP20-29 | Ach18g165371 | 4.99 | 22.60 | 198 | Chr18: 8741760.. 8742825(+) | 细胞质Cytoplasmic |
| AcHSP20-30 | Ach19g138461.2 | 5.08 | 13.25 | 114 | Chr19: 7272801.. 7273145(+) | 细胞质Cytoplasmic |
| AcHSP20-31 | Ach22g015991.2 | 7.61 | 26.63 | 241 | Chr22: 8846412.. 8851047(-) | 叶绿体Chloroplast |
| AcHSP20-32 | Ach23g121711.2 | 5.37 | 22.34 | 199 | Chr23: 4320454.. 4321535(-) | 细胞质Cytoplasmic |
| AcHSP20-33 | Ach23g411051.2 | 9.11 | 33.06 | 300 | Chr23: 19087591.. 19088582(+) | 叶绿体Chloroplast |
| AcHSP20-34 | Ach29g196461.2 | 4.59 | 9.84 | 85 | Chr29: 3710817.. 3711074(+) | 叶绿体Chloroplast |
表2 AcHSP20s蛋白质理化性质
Table 2 Physicochemical properties of AcHSP20s proteins in kiwifruit
| 基因 Gene | 基因组登录号 Gene ID | 等电点 pI | 分子量 Molecular weight/kD | 氨基酸数Number of amino acids | 染色体定位及基因方向Chromosome localization and gene orientation | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|
| AcHSP20-1 | Ach00g218421.2 | 6.57 | 10.16 | 88 | Chr00: 54153215.. 54154904(-) | 细胞质Cytoplasmic |
| AcHSP20-2 | Ach00g239181 | 7.86 | 16.14 | 143 | Chr00: 50739806.. 50740562(-) | 过氧化物酶体Peroxisomal |
| AcHSP20-3 | Ach00g329621 | 5.59 | 23.28 | 208 | Chr00: 90841963.. 90843153(-) | 叶绿体Chloroplast |
| AcHSP20-4 | Ach00g479741.2 | 10.32 | 9.93 | 86 | Chr00: 135211617.. 135211877(+) | 细胞质Cytoplasmic |
| AcHSP20-5 | Ach00g479761.2 | 9.14 | 40.18 | 371 | Chr00: 145805825.. 145808080(-) | 高尔基体Golgi apparatus |
| AcHSP20-6 | Ach02g036831 | 7.8 | 16.56 | 152 | Chr02: 4423490.. 4423948(-) | 细胞质Cytoplasmic |
| AcHSP20-7 | Ach02g046741.2 | 6.21 | 25.55 | 225 | Chr02: 516872.. 518080(-) | 叶绿体Chloroplast |
| AcHSP20-8 | Ach02g223291 | 8.86 | 17.61 | 153 | Chr02: 4090770.. 4091231(-) | 细胞质Cytoplasmic |
| AcHSP20-9 | Ach03g045661 | 7.63 | 25.69 | 232 | Chr03: 6797048.. 6798489(-) | 叶绿体Chloroplast |
| AcHSP20-10 | Ach03g451751.2 | 9.15 | 11.12 | 96 | Chr03: 1045510.. 1045800(-) | 叶绿体Chloroplast |
| AcHSP20-11 | Ach04g161211.2 | 6.45 | 24.77 | 215 | Chr04: 6179073.. 6180486(-) | 细胞质Cytoplasmic |
| AcHSP20-12 | Ach06g418911.2 | 6.32 | 16.60 | 152 | Chr06: 2925504.. 2925962(-) | 细胞质Cytoplasmic |
| AcHSP20-13 | Ach10g027441.2 | 5.4 | 20.33 | 181 | Chr10: 4989172.. 4989717(-) | 细胞质Cytoplasmic |
| AcHSP20-14 | Ach10g027451 | 7.77 | 25.72 | 228 | Chr10: 4973842.. 4974528(-) | 细胞质Cytoplasmic |
| AcHSP20-15 | Ach10g027461.2 | 6.34 | 24.95 | 221 | Chr10: 4984511.. 4985176(+) | 叶绿体Chloroplast |
| AcHSP20-16 | Ach10g143161 | 9.3 | 25.78 | 228 | Chr10: 11814664.. 11815350(+) | 细胞质Cytoplasmic |
| AcHSP20-17 | Ach11g263431.2 | 5.05 | 21.72 | 196 | Chr11: 8776984.. 8777574(-) | 细胞质Cytoplasmic |
| AcHSP20-18 | Ach11g425231.2 | 4.43 | 10.59 | 90 | Chr11: 12263315.. 12263587(-) | 细胞质Cytoplasmic |
| AcHSP20-19 | Ach12g058351.2 | 8.69 | 17.21 | 152 | Chr12: 6955029.. 6956940(+) | 细胞核Nucleus |
| AcHSP20-20 | Ach13g308811 | 5.94 | 33.07 | 297 | Chr13: 1116598.. 1117580(+) | 叶绿体Chloroplast |
| AcHSP20-21 | Ach13g350901 | 5.68 | 21.90 | 195 | Chr13: 14397259.. 14398212(+) | 叶绿体Chloroplast |
| AcHSP20-22 | Ach13g383281 | 6.31 | 16.61 | 152 | Chr13: 11985660.. 11986118(-) | 细胞质Cytoplasmic |
| AcHSP20-23 | Ach13g472651.2 | 5.94 | 33.07 | 297 | Chr13: 1497079.. 1498061(-) | 叶绿体Chloroplast |
| AcHSP20-24 | Ach14g205801.2 | 6.87 | 24.06 | 216 | Chr14: 9422059.. 9423375(+) | 叶绿体Chloroplast |
| AcHSP20-25 | Ach14g205811.2 | 8.86 | 36.76 | 329 | Chr14: 9424474.. 9428179(+) | 叶绿体Chloroplast |
| AcHSP20-26 | Ach15g376651 | 5.67 | 22.90 | 202 | Chr15: 4758959.. 4759567(-) | 细胞质Cytoplasmic |
| AcHSP20-27 | Ach16g309411.2 | 7.64 | 26.14 | 231 | Chr16: 8508810.. 8510240(-) | 叶绿体Chloroplast |
| AcHSP20-28 | Ach16g331111 | 8.83 | 17.36 | 153 | Chr16: 10507526.. 10508507(+) | 细胞质Cytoplasmic |
| AcHSP20-29 | Ach18g165371 | 4.99 | 22.60 | 198 | Chr18: 8741760.. 8742825(+) | 细胞质Cytoplasmic |
| AcHSP20-30 | Ach19g138461.2 | 5.08 | 13.25 | 114 | Chr19: 7272801.. 7273145(+) | 细胞质Cytoplasmic |
| AcHSP20-31 | Ach22g015991.2 | 7.61 | 26.63 | 241 | Chr22: 8846412.. 8851047(-) | 叶绿体Chloroplast |
| AcHSP20-32 | Ach23g121711.2 | 5.37 | 22.34 | 199 | Chr23: 4320454.. 4321535(-) | 细胞质Cytoplasmic |
| AcHSP20-33 | Ach23g411051.2 | 9.11 | 33.06 | 300 | Chr23: 19087591.. 19088582(+) | 叶绿体Chloroplast |
| AcHSP20-34 | Ach29g196461.2 | 4.59 | 9.84 | 85 | Chr29: 3710817.. 3711074(+) | 叶绿体Chloroplast |

图2 AcHSP20s的染色体定位和共线性分析 A:猕猴桃34个AcHSP20s的染色体定位;B:猕猴桃34个AcHSP20s基因的共线性分析;C:猕猴桃34个AcHSP20s和拟南芥的19个AtHSP20s的种间共线性分析
Fig. 2 Chromosomal localization and synteny analysis of AcHSP20s A: Chromosomal localizations of 34 AcHSP20s in kiwifruit. B: Synteny analysis of 34 AcHSP20s in kiwifruit. C: Synteny analysis between 34 AcHSP20s in kiwifruit and 19 AtHSP20s in Arabidopsis
| 基因对Gene pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| AcHSP20-7/AcHSP20-10 | 0.231 715 656 | 0.423 821 440 | 0.546 729 433 |
| AcHSP20-7/AcHSP20-9 | 0.209 150 940 | 0.704 599 321 | 0.296 836 704 |
| AcHSP20-10/AcHSP20-9 | 0.322 818 372 | 0.919 187 813 | 0.351 199 578 |
| AcHSP20-18/AcHSP20-27 | 0.102 831 910 | 0.278 905 623 | 0.368 697 875 |
| AcHSP20-20/AcHSP20-33 | 0.104 764 648 | 0.138 864 084 | 0.754 440 205 |
| AcHSP20-23/AcHSP20-33 | 0.104 764 648 | 0.138 864 084 | 0.754 440 205 |
| AcHSP20-21/AcHSP20-34 | 0.092 066 167 | 0.397 378 202 | 0.231 683 988 |
| AcHSP20-30/AcHSP20-32 | 0.239 953 460 | 0.526 314 021 | 0.455 913 107 |
表3 AcHSP20s的Ka/Ks值
Table 3 Ka/Ks value of AcHSP20s
| 基因对Gene pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| AcHSP20-7/AcHSP20-10 | 0.231 715 656 | 0.423 821 440 | 0.546 729 433 |
| AcHSP20-7/AcHSP20-9 | 0.209 150 940 | 0.704 599 321 | 0.296 836 704 |
| AcHSP20-10/AcHSP20-9 | 0.322 818 372 | 0.919 187 813 | 0.351 199 578 |
| AcHSP20-18/AcHSP20-27 | 0.102 831 910 | 0.278 905 623 | 0.368 697 875 |
| AcHSP20-20/AcHSP20-33 | 0.104 764 648 | 0.138 864 084 | 0.754 440 205 |
| AcHSP20-23/AcHSP20-33 | 0.104 764 648 | 0.138 864 084 | 0.754 440 205 |
| AcHSP20-21/AcHSP20-34 | 0.092 066 167 | 0.397 378 202 | 0.231 683 988 |
| AcHSP20-30/AcHSP20-32 | 0.239 953 460 | 0.526 314 021 | 0.455 913 107 |

图4 AcHSP20s启动子顺式作用元件分析 A:猕猴桃34个AcHSP20基因启动子顺式作用元件具体位置;B:猕猴桃34个AcHSP20基因启动子顺式作用元件数量
Fig. 4 Investigation of cis-acting elements in the promoters of AcHSP20s A: The locations of cis-acting elements of 34 AcHSP20s in kiwifruit. B: Numbers of 34 AcHSP20s cis-acting elements in kiwifruit
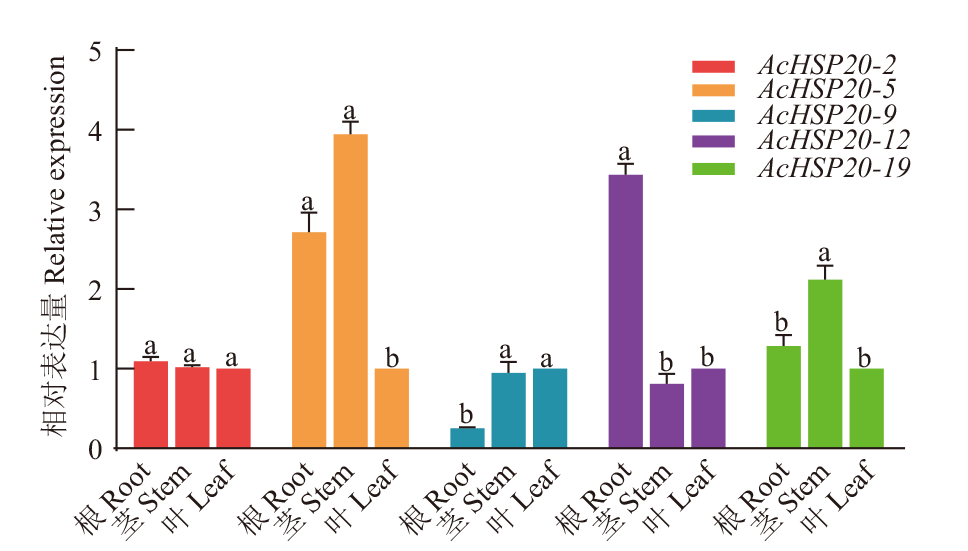
图5 AcHSP20s在猕猴桃中的表达模式 3次生物学重复和3次技术重复,标准误差显示在柱形图上,小写字母表示不同样品间的差异(P<0.05)。下同
Fig. 5 Expression profiles of AcHSP20s in kiwifruit There are 3 biological and technical replicates, the standard error is on the bar graph, and lowercase letters indicate the difference among different samples. The same below
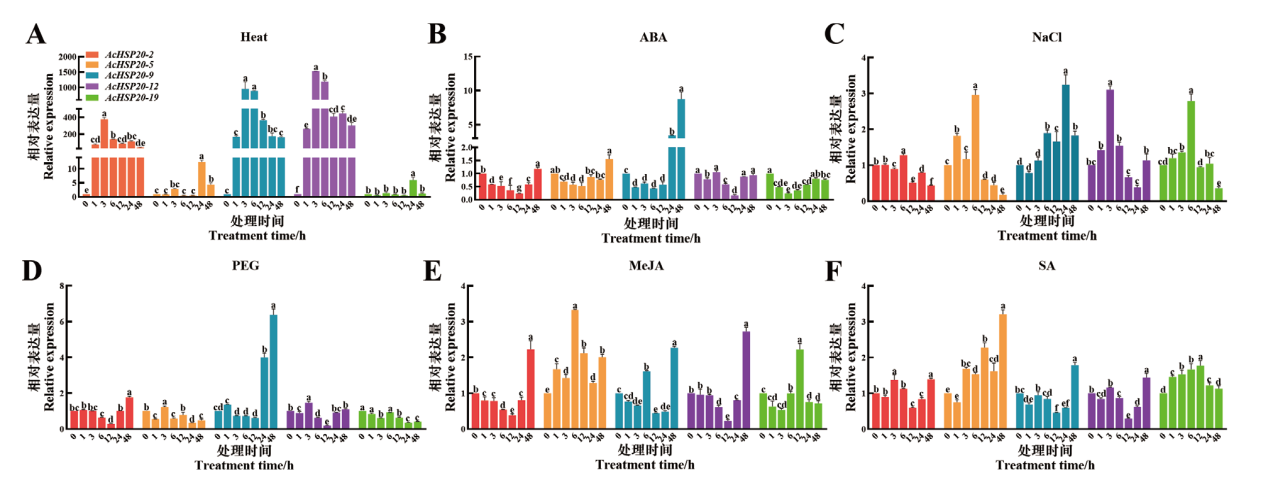
图6 AcHSP20s在非生物胁迫、植物激素和植物生长调节剂处理下的表达情况
Fig. 6 Relative expressions of AcHSP20s under abiotic stress, phytohormone and plant growth regulator treatments
| [1] |
Herman DJ, Knowles LO, Knowles NR. Heat stress affects carbohydrate metabolism during cold-induced sweetening of potato(Solanum tuberosum L.)[J]. Planta, 2017, 245(3): 563-582.
doi: 10.1007/s00425-016-2626-z pmid: 27904974 |
| [2] |
Giorno F, Wolters-Arts M, Grillo S, et al. Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers[J]. J Exp Bot, 2010, 61(2): 453-462.
doi: 10.1093/jxb/erp316 pmid: 19854799 |
| [3] |
Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants[J]. Curr Opin Plant Biol, 2011, 14(3): 290-295.
doi: 10.1016/j.pbi.2011.02.001 pmid: 21377404 |
| [4] |
Wang N, Guo TL, Sun X, et al. Functions of two Malus hupehensis(Pamp.) Rehd. YTPs(MhYTP1 and MhYTP2)in biotic- and abiotic-stress responses[J]. Plant Sci, 2017, 261: 18-27.
doi: S0168-9452(17)30238-8 pmid: 28554690 |
| [5] | Suzuki N. Temperature stress and responses in plants[J]. Int J Mol Sci, 2019, 20(8): 2001. |
| [6] |
Yang GD, Yu ZP, Gao L, et al. SnRK2s at the crossroads of growth and stress responses[J]. Trends Plant Sci, 2019, 24(8): 672-676.
doi: S1360-1385(19)30139-6 pmid: 31255544 |
| [7] |
Tang XL, Mu XM, Shao HB, et al. Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology[J]. Crit Rev Biotechnol, 2015, 35(4): 425-437.
doi: 10.3109/07388551.2014.889080 pmid: 24738851 |
| [8] | Tang XL, Shao HB, Jiang FD, et al. Molecular cloning and functional analyses of the salt-responsive gene KvHSP70 from Kosteletzkya virginica[J]. Land Degrad Dev, 2020, 31(6): 773-782. |
| [9] | Wu JT, Gao T, Hu JN, et al. Research advances in function and regulation mechanisms of plant small heat shock proteins(sHSPs)under environmental stresses[J]. Sci Total Environ, 2022, 825: 154054. |
| [10] |
Jacob P, Hirt H, Bendahmane A. The heat-shock protein/chaperone network and multiple stress resistance[J]. Plant Biotechnol J, 2017, 15(4): 405-414.
doi: 10.1111/pbi.12659 pmid: 27860233 |
| [11] | Zhang KM, Ezemaduka AN, Wang Z, et al. A novel mechanism for small heat shock proteins to function as molecular chaperones[J]. Sci Rep, 2015, 5(5): 8811-8819. |
| [12] | Haq S U, Khan A, Ali M, et al. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses[J]. Int J Mol Sci, 2019, 20(21): 5321. |
| [13] | Tian C, Zhang ZY, Huang Y, et al. Functional characterization of the Pinellia ternata cytoplasmic class II small heat shock protein gene PtsHSP17.2 via promoter analysis and overexpression in tobacco[J]. Plant Physiol Biochem, 2022, 177: 1-9. |
| [14] | Feng XH, Zhang HX, Ali M, et al. A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper(Capsicum annuum L.)[J]. Plant Physiol Biochem, 2019, 142(142): 151-162. |
| [15] | Waters ER. The evolution, function, structure and expression of the plant sHSPs[J]. J Exp Bot, 2013, 64(2): 391-403. |
| [16] | Waters ER, Vierling E. Plant small heat shock proteins-evolutionary and functional diversity[J]. New Phytol, 2020, 227(1): 24-37. |
| [17] |
Haslbeck M, Franzmann T, Weinfurtner D, et al. Some like it hot: the structure and function of small heat-shock proteins[J]. Nat Struct Mol Biol, 2005, 12(10): 842-846.
doi: 10.1038/nsmb993 pmid: 16205709 |
| [18] | El-Gebali S, Mistry J, Bateman A, et al. The Pfam protein families database in 2019[J]. Nucleic Acids Res, 2019, 47(D1): D427-D432. |
| [19] |
Ma W, Guan XY, Li J, et al. Mitochondrial small heat shock protein mediates seed germination via thermal sensing[J]. Proc Natl Acad Sci USA, 2019, 116(10): 4716-4721.
doi: 10.1073/pnas.1815790116 pmid: 30765516 |
| [20] |
Rutsdottir G, Härmark J, Weide Y, et al. Structural model of dodecameric heat-shock protein Hsp21: Flexible N-terminal arms interact with client proteins while C-terminal tails maintain the dodecamer and chaperone activity[J]. J Biol Chem, 2017, 292(19): 8103-8121.
doi: 10.1074/jbc.M116.766816 pmid: 28325834 |
| [21] |
Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation[J]. EMBO J, 2008, 27(2): 328-335.
doi: 10.1038/sj.emboj.7601970 pmid: 18216875 |
| [22] | Yang ML, Zhang YX, Zhang HH, et al. Identification of MsHsp20 gene family in malus sieversii and functional characterization of MsHsp16.9 in heat tolerance[J]. Front Plant Sci, 2017, 8: 1761-1778. |
| [23] | Sewelam N, Kazan K, Hüdig M, et al. The AtHSP17.4C1 gene expression is mediated by diverse signals that link biotic and abiotic stress factors with ROS and can be a useful molecular marker for oxidative stress[J]. Int J Mol Sci, 2019, 20(13): 3201-3218. |
| [24] | Mu CJ, Zhang SJ, Yu GZ, et al. Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses[J]. PLoS One, 2013, 8(12): e82264-e82273. |
| [25] | Zhang L, Gao YK, Pan HT, et al. Cloning and characterisation of a Primula heat shock protein gene, PfHSP17.1, which confers heat, salt and drought tolerance in transgenic Arabidopsis thaliana[J]. Acta Physiol Plant, 2013, 35(11): 3191-3200. |
| [26] | Sun XB, Sun CY, Li ZG, et al. AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress[J]. Plant Cell Environ, 2016, 39(6): 1320-1337. |
| [27] | Zhang N, Zhao HY, Shi JW, et al. Functional characterization of class I SlHSP17.7 gene responsible for tomato cold-stress tolerance[J]. Plant Sci, 2020, 298: 110568. |
| [28] | Do VG, Lee Y, Kweon H, et al. Light induces carotenoid biosynthesis-related gene expression, accumulation of pigment content, and expression of the small heat shock protein in apple fruit[J]. Int J Mol Sci, 2022, 23(11): 6153. |
| [29] |
Kaur H, Petla BP, Kamble NU, et al. Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress[J]. Front Plant Sci, 2015, 6: 713.
doi: 10.3389/fpls.2015.00713 pmid: 26442027 |
| [30] |
Kuang J, Liu JZ, Mei J, et al. A class II small heat shock protein OsHsp18.0 plays positive roles in both biotic and abiotic defense responses in rice[J]. Sci Rep, 2017, 7(1): 11333.
doi: 10.1038/s41598-017-11882-x pmid: 28900229 |
| [31] |
Abassi S, Wang H, Ponmani T, et al. Small heat shock protein genes of the green algae Closterium ehrenbergii: Cloning and differential expression under heat and heavy metal stresses[J]. Environ Toxicol, 2019, 34(9): 1013-1024.
doi: 10.1002/tox.22772 pmid: 31095847 |
| [32] | Li ZY, Long RC, Zhang TJ, et al. Molecular cloning and characterization of the MsHSP17.7 gene from Medicago sativa L.[J]. Mol Biol Rep, 2016, 43(8): 815-826. |
| [33] | Li XW, Li JQ, Djaja DS. New synonyms in Actinidiaceae from China[J]. J Syst Evol, 2007, 45(5): 633-660. |
| [34] | 钟彩虹, 黄文俊, 李大卫, 等. 世界猕猴桃产业发展及鲜果贸易动态分析[J]. 中国果树, 2021, 7(7): 101-108. |
| Zhong CH, Huang WJ, Li DW, et al. Dynamic analysis of global kiwifruit industry development and fresh fruit trade[J]. China Fruits, 2021, 7(7): 101-108. | |
| [35] | Hua YG, Liu Q, Zhai YF, et al. Genome-wide analysis of the HSP20 gene family and its response to heat and drought stress in Coix(Coix lacryma-jobi L.)[J]. BMC Genomics, 2023, 24(1): 478-494. |
| [36] | Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains(Acd proteins)[J]. Cell Stress Chaperones, 2001, 6(3): 225-237. |
| [37] |
Ouyang YD, Chen JJ, Xie WB, et al. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice[J]. Plant Mol Biol, 2009, 70(3): 341-357.
doi: 10.1007/s11103-009-9477-y pmid: 19277876 |
| [38] |
Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms[J]. Mol Biol Evol, 2001, 18(9): 1611-1630.
doi: 10.1093/oxfordjournals.molbev.a003951 pmid: 11504843 |
| [39] | Huang LJ, Cheng GX, Khan A, et al. CaHSP16.4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance[J]. Protoplasma, 2019, 256(1): 39-51. |
| [40] | He YJ, Yao YX, Li LL, et al. A heat-shock 20 protein isolated from watermelon(ClHSP22.8)negatively regulates the response of Arabidopsis to salt stress via multiple signaling pathways[J]. PeerJ, 2021, 9: e10524. |
| [1] | 李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202. |
| [2] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [3] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [4] | 张清兰, 张亚冉, 鞠志花, 王秀革, 肖遥, 王金鹏, 魏晓超, 高亚平, 白福恒, 王洪程. 牛TARDBP基因核心启动子鉴定与转录调控分析[J]. 生物技术通报, 2024, 40(4): 306-318. |
| [5] | 陈晓松, 刘超杰, 郑佳, 乔宗伟, 罗惠波, 邹伟. TMT定量蛋白质组学解析Rummeliibacillus suwonensis 3B-1 生长及己酸代谢机制[J]. 生物技术通报, 2024, 40(3): 135-145. |
| [6] | 杨伟杰, 杨周林, 朱浩东, 魏煜, 刘君, 刘训. 地衣素合成酶关键模块 LchAD 蛋白的性质和功能研究[J]. 生物技术通报, 2024, 40(3): 322-332. |
| [7] | 龚丽丽, 余花, 杨杰, 陈天池, 赵双滢, 吴月燕. 葡萄CYP707A基因家族的鉴定及对果实成熟的功能验证[J]. 生物技术通报, 2024, 40(2): 160-171. |
| [8] | 路喻丹, 刘晓驰, 冯新, 陈桂信, 陈义挺. 猕猴桃BBX基因家族成员鉴定与转录特征分析[J]. 生物技术通报, 2024, 40(2): 172-182. |
| [9] | 苑馨予, 钟彩虹, 张龙, 郑浩, 李吉涛, 张琼. 猕猴桃AcMYB88的鉴定及功能研究[J]. 生物技术通报, 2024, 40(2): 183-196. |
| [10] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [11] | 李敬蕊, 王育博, 解紫薇, 李畅, 吴晓蕾, 宫彬彬, 高洪波. 甜瓜PIN基因家族的鉴定及高温胁迫表达分析[J]. 生物技术通报, 2023, 39(5): 192-204. |
| [12] | 赖瑞联, 冯新, 高敏霞, 路喻丹, 刘晓驰, 吴如健, 陈义挺. 猕猴桃过氧化氢酶基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2023, 39(4): 136-147. |
| [13] | 郭三保, 宋美玲, 李灵心, 尧子钊, 桂明明, 黄胜和. 斑地锦查尔酮合酶基因及启动子的克隆与分析[J]. 生物技术通报, 2023, 39(4): 148-156. |
| [14] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [15] | 杨岚, 张晨曦, 樊学伟, 王阳光, 王春秀, 李文婷. 鸡 BMP15 基因克隆、表达模式及启动子活性分析[J]. 生物技术通报, 2023, 39(4): 304-312. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||