








生物技术通报 ›› 2024, Vol. 40 ›› Issue (3): 135-145.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0862
陈晓松1( ), 刘超杰1, 郑佳2,3, 乔宗伟2,3, 罗惠波1, 邹伟1,2(
), 刘超杰1, 郑佳2,3, 乔宗伟2,3, 罗惠波1, 邹伟1,2( )
)
收稿日期:2023-09-06
出版日期:2024-03-26
发布日期:2024-04-08
通讯作者:
邹伟,男,博士,副教授,研究方向:白酒微生物;E-mail: weizou@suse.edu.cn作者简介:陈晓松,男,硕士研究生,研究方向:发酵工程;E-mail: xschenyx@qq.com
基金资助:
CHEN Xiao-song1( ), LIU Chao-jie1, ZHENG Jia2,3, QIAO Zong-wei2,3, LUO Hui-bo1, ZOU Wei1,2(
), LIU Chao-jie1, ZHENG Jia2,3, QIAO Zong-wei2,3, LUO Hui-bo1, ZOU Wei1,2( )
)
Received:2023-09-06
Published:2024-03-26
Online:2024-04-08
摘要:
【目的】 从蛋白水平阐明水源拉梅尔芽孢杆菌(Rummeliibacillus suwonensis)的生长及己酸代谢机理,为水源拉梅尔芽孢杆菌的基因工程改造提供一定技术基础。【方法】 以R. suwonensis 3B-1为研究对象,应用串联质谱标签(tandem mass tags, TMT)蛋白组学技术对该菌在好氧与厌氧条件下的差异表达蛋白(differentially expressed proteins, DEPs)进行挖掘,并对鉴定到的DEPs进行亚细胞定位、GO功能富集、KEGG信号通路注释、蛋白相互作用等生物信息学分析。【结果】 从比较组中共鉴定获得810个DEPs,其中上调蛋白423个,下调蛋白387个,亚细胞定位到6个条目上,主要涉及细胞质蛋白,细胞膜蛋白和细胞壁等蛋白。GO功能富集分析结果显示,肽的生物合成、翻译和肽代谢过程等生物学过程;核糖体的结构组成和结构分子活性等分子功能;核糖体和核糖核蛋白复合物等细胞组分发生了显著变化。810个DEPs 注释到113条KEGG信号通路,主要涉及辅因子生物合成,双组分系统,磷酸戊糖代谢,糖酵解/糖异生,以及氧化磷酸化等信号通路。苯丙氨酸-tRNA连接酶β亚基和核黄素生物合成蛋白RibD在蛋白互作网络中关联度最高。【结论】 厌氧条件下,糖酵解途径中丙酮酸脱氢酶和丙酮酸激酶表达下调,氨基酸代谢和生物素蛋白连接酶等辅因子相关蛋白表达均呈现下调,表明该菌适合在好氧环境中生长。己酸合成方面,酰基辅酶A硫酯酶的表达量显著上调,同时,糖酵解/糖异生途径、三羧酸循环和磷酸戊糖途径为己酸合成提供了充足的前体物质和还原当量,共同促进了己酸合成。
陈晓松, 刘超杰, 郑佳, 乔宗伟, 罗惠波, 邹伟. TMT定量蛋白质组学解析Rummeliibacillus suwonensis 3B-1 生长及己酸代谢机制[J]. 生物技术通报, 2024, 40(3): 135-145.
CHEN Xiao-song, LIU Chao-jie, ZHENG Jia, QIAO Zong-wei, LUO Hui-bo, ZOU Wei. Analyzing the Growth and Caproic Acid Metabolism Mechanism of Rummeliibacillus suwonensis 3B-1 by Tandem Mass Tag-based Quantitative Proteomics[J]. Biotechnology Bulletin, 2024, 40(3): 135-145.
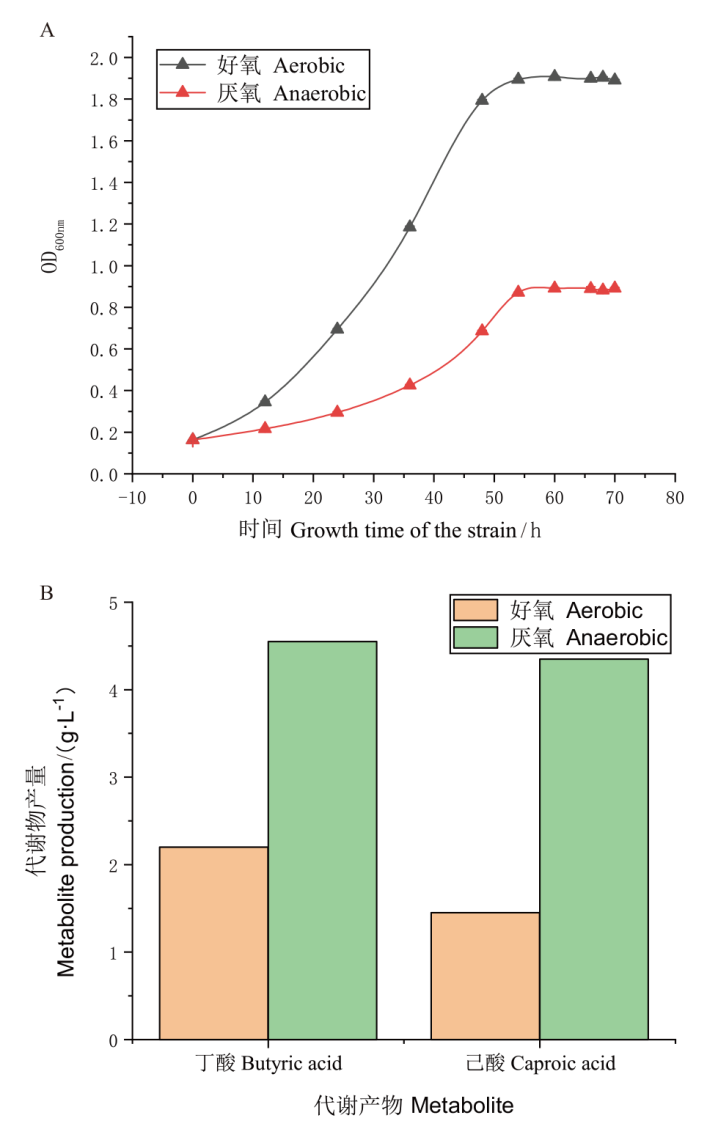
图1 3B-1好氧和厌氧条件生长及代谢情况 A:好氧和厌氧条件下3B-1生长曲线图;B:好氧和厌氧条件下3B-1丁酸、乙酸产量图
Fig. 1 Growth and metabolism of 3B-1 under aerobic and anaerobic conditions A : 3B-1 growth curves under aerobic and anaerobic conditions. B : Butyric acid and caproic acid production of 3B-1 under aerobic and anaerobic conditions
| 样品名称 Sample | 编号 Sample number | 蛋白浓度 Protein concentration/(μg·μL-1) | 蛋白总量 Total protein/μg |
|---|---|---|---|
| RH1 | 1 | 1.475 | 885 |
| RH2 | 2 | 3.197 | 1 918.2 |
| RH3 | 3 | 1.647 | 988.2 |
| RY1 | 4 | 1.268 | 760.8 |
| RY2 | 5 | 1.954 | 1 172.4 |
| RY3 | 6 | 1.608 | 964.8 |
表1 BCA蛋白定量结果
Table 1 BCA protein quantification results
| 样品名称 Sample | 编号 Sample number | 蛋白浓度 Protein concentration/(μg·μL-1) | 蛋白总量 Total protein/μg |
|---|---|---|---|
| RH1 | 1 | 1.475 | 885 |
| RH2 | 2 | 3.197 | 1 918.2 |
| RH3 | 3 | 1.647 | 988.2 |
| RY1 | 4 | 1.268 | 760.8 |
| RY2 | 5 | 1.954 | 1 172.4 |
| RY3 | 6 | 1.608 | 964.8 |
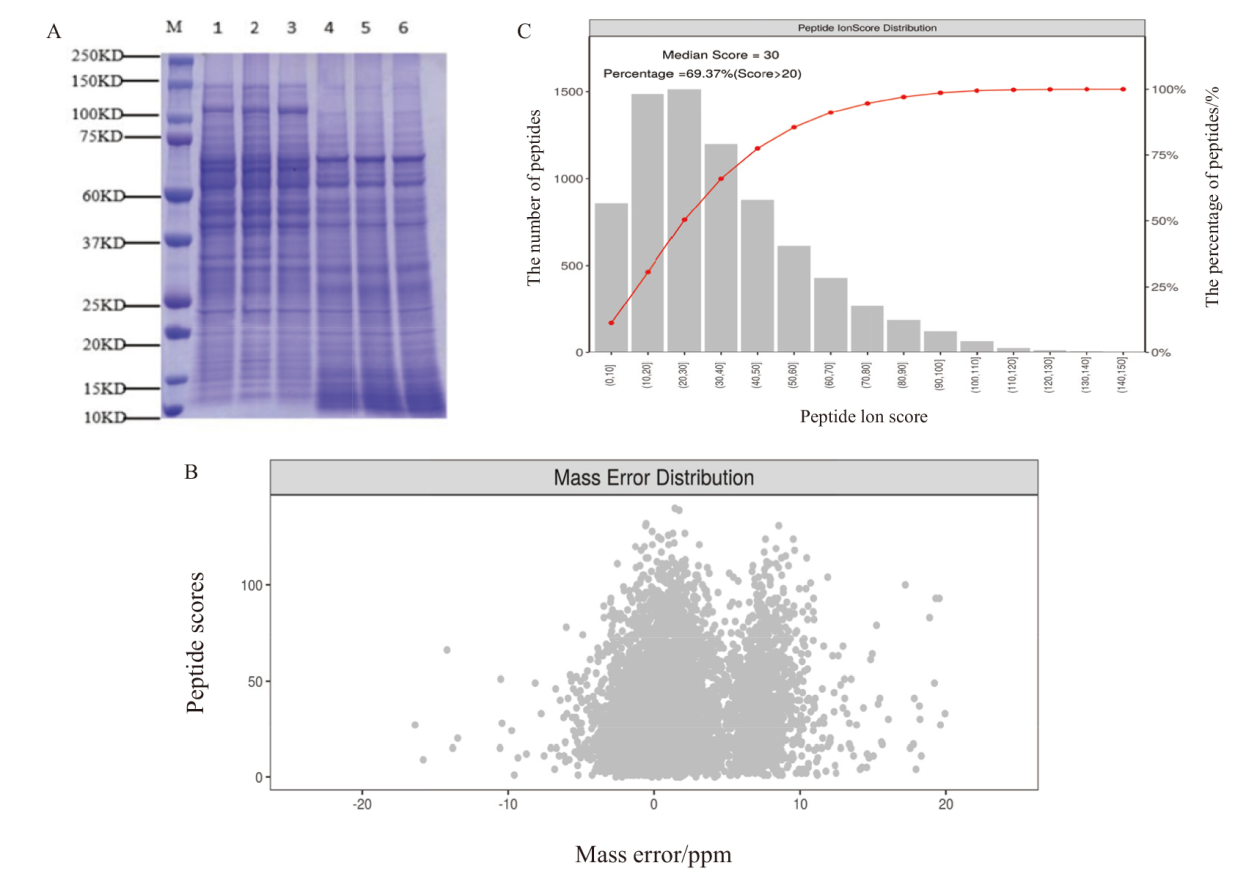
图2 蛋白样品质量控制 A:蛋白样品SDS-PAGE图;B:肽段离子质量偏差分布图;C:肽段离子得分分布图
Fig. 2 Quality control of protein sample A: SDS-PAGE of protein samples. B: Distribution plot of peptide ion mass deviations. C: Distribution plot of peptide ion scores
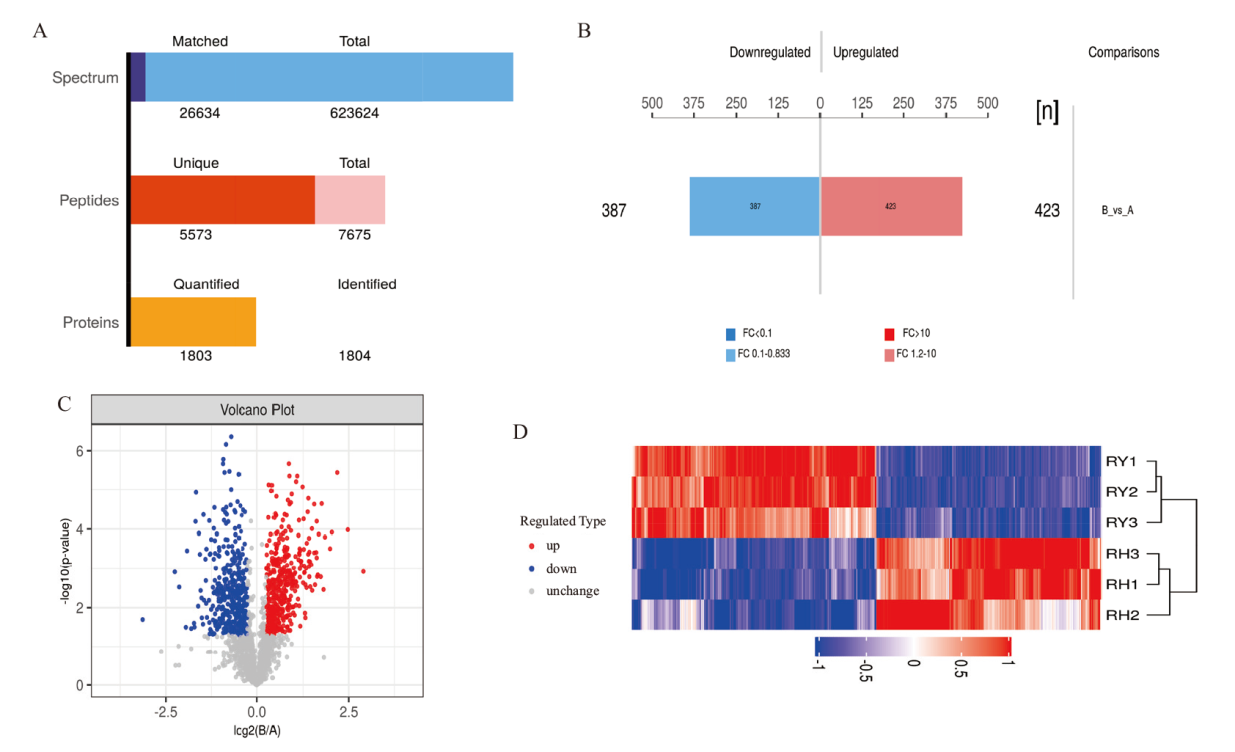
图3 蛋白鉴定和差异表达结果 A:蛋白质的鉴定与定量结果图;B:蛋白质定量差异结果柱状图;C:比较组差异显著性火山图;D:比较组DEPs聚类分析图
Fig. 3 Results of protein identification and differential expression A: Result graph of protein identification and quantification. B: Bar graph of protein quantification differences. C: Volcano plot of significant differences in the comparative group. D: Analysis graph of clustering of DEPs in the comparative group
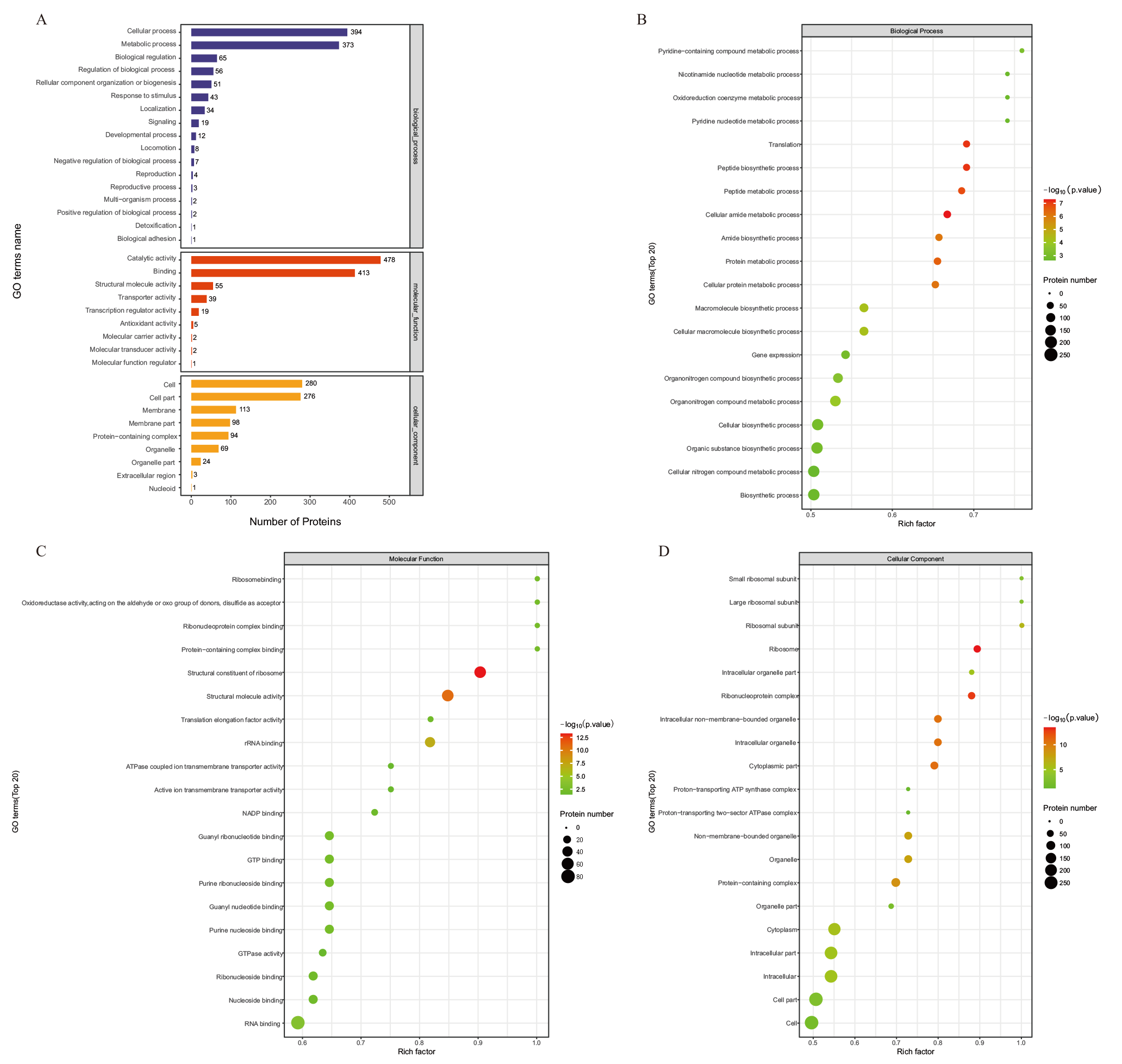
图4 比较组DEPs的GO注释图 A:比较组DEPs的GO注释图;B:比较组DEPs的生物过程分类下GO功能富集气泡图;C:比较组DEPs的分子功能分类下GO功能富集气泡图D:比较组DEPs的细胞组分分类下GO功能富集气泡图
Fig. 4 GO annotation map of DEPs in comparative group A : GO annotation map of DEPs in the comparative group. B : GO functional enrichment bubble diagram under the biological process classification of DEPs in the comparative group. C: GO functional enrichment bubble diagram under the molecular function classification of DEPs in the comparative group. D : GO functional enrichment bubble diagram under the cell component classification of DEPs in the comparative group
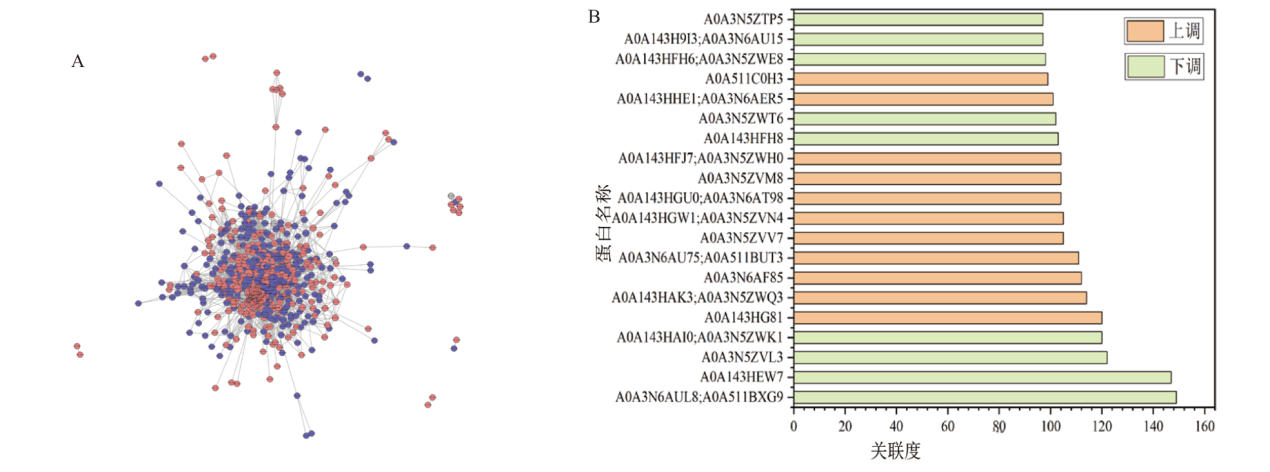
图6 比较组DEPs互作网络分析结果 A:比较组DEPs网络互作图; B: DEPs互作网络中关联度最高的20个蛋白表达情况
Fig. 6 Results of the interaction network analysis of DEP in the comparative groups A: Network interaction graph of DEPs in the comparative groups. B: Expression patterns of the top 20 proteins with the highest correlation in the DEPs interaction network
| 蛋白编号Protein | 基因名称Gene name | 蛋白描述Annotation |
|---|---|---|
| A0A3N6AUL8;A0A511BXG9 | pheT | Phenylalanine--tRNA ligase beta subunit |
| A0A143HEW7 | — | Riboflavin biosynthesis protein RibD |
| A0A3N5ZVL3 | rpoA | DNA-directed RNA polymerase subunit alpha |
| A0A143HAI0;A0A3N5ZWK1 | lepA | Elongation factor 4 |
| A0A143HG81 | tuf | Elongation factor Tu |
| A0A143HAK3;A0A3N5ZWQ3 | rpsB | 30S ribosomal protein S2 |
| A0A3N6AF85 | rplB | 50S ribosomal protein L2 |
| A0A3N6AU75;A0A511BUT3 | infB | Translation initiation factor IF-2 |
| A0A3N5ZVV7 | rplE | 50S ribosomal protein L5 |
| A0A143HGW1;A0A3N5ZVN4 | fusA | Elongation factor G |
| A0A143HGU0;A0A3N6AT98 | rpsE | 30S ribosomal protein S5 |
| A0A3N5ZVM8 | rpsC | 30S ribosomal protein S3 |
| A0A143HFJ7;A0A3N5ZWH0 | rpsG | 30S ribosomal protein S7 |
| A0A143HFH8 | rplD | 50S ribosomal protein L4 |
| A0A3N5ZWT6 | cinA | Putative competence-damage inducible |
| A0A143HHE1;A0A3N6AER5 | atpA | ATP synthase subunit alpha |
| A0A511C0H3 | rplR | 50S ribosomal protein L18 |
| A0A143HFH6;A0A3N5ZWE8 | rpsK | 30S ribosomal protein S11 |
| A0A143H9I3;A0A3N6AU15 | ftsY | Signal recognition particle receptor FtsY |
| A0A3N5ZTP5 | birA | Bifunctional ligase/repressor BirA |
表2 比较组蛋白互作网络中关联度最高的20个蛋白质
Table 2 Top 20 proteins with the highest correlation in the interaction network of DEPs
| 蛋白编号Protein | 基因名称Gene name | 蛋白描述Annotation |
|---|---|---|
| A0A3N6AUL8;A0A511BXG9 | pheT | Phenylalanine--tRNA ligase beta subunit |
| A0A143HEW7 | — | Riboflavin biosynthesis protein RibD |
| A0A3N5ZVL3 | rpoA | DNA-directed RNA polymerase subunit alpha |
| A0A143HAI0;A0A3N5ZWK1 | lepA | Elongation factor 4 |
| A0A143HG81 | tuf | Elongation factor Tu |
| A0A143HAK3;A0A3N5ZWQ3 | rpsB | 30S ribosomal protein S2 |
| A0A3N6AF85 | rplB | 50S ribosomal protein L2 |
| A0A3N6AU75;A0A511BUT3 | infB | Translation initiation factor IF-2 |
| A0A3N5ZVV7 | rplE | 50S ribosomal protein L5 |
| A0A143HGW1;A0A3N5ZVN4 | fusA | Elongation factor G |
| A0A143HGU0;A0A3N6AT98 | rpsE | 30S ribosomal protein S5 |
| A0A3N5ZVM8 | rpsC | 30S ribosomal protein S3 |
| A0A143HFJ7;A0A3N5ZWH0 | rpsG | 30S ribosomal protein S7 |
| A0A143HFH8 | rplD | 50S ribosomal protein L4 |
| A0A3N5ZWT6 | cinA | Putative competence-damage inducible |
| A0A143HHE1;A0A3N6AER5 | atpA | ATP synthase subunit alpha |
| A0A511C0H3 | rplR | 50S ribosomal protein L18 |
| A0A143HFH6;A0A3N5ZWE8 | rpsK | 30S ribosomal protein S11 |
| A0A143H9I3;A0A3N6AU15 | ftsY | Signal recognition particle receptor FtsY |
| A0A3N5ZTP5 | birA | Bifunctional ligase/repressor BirA |
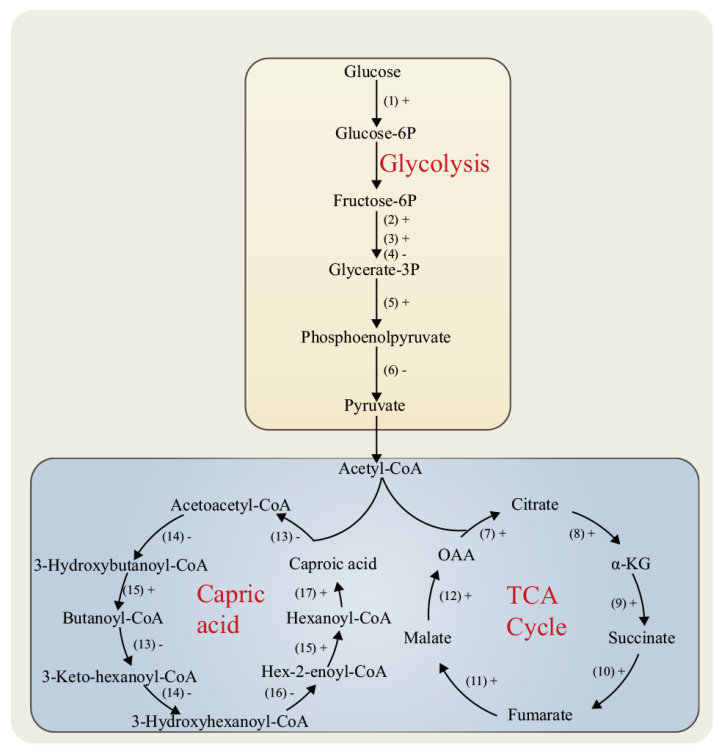
图7 DEPs在中心代谢和己酸代谢途径中的分布 “+”表示该酶表达量上调,“-”表示该酶表达量下调。(1):己糖激酶;(2):ATP依赖的6-磷酸果糖激酶;(3):磷酸甘油酸激酶;(4):果糖二磷酸醛缩酶;(5):2 -磷酸甘油酸脱水酶;(6):丙酮酸激酶;(7):柠檬酸合酶;(8):异柠檬酸脱氢酶;(9):α-酮戊二酸脱氢酶复合体;(10):琥珀酸脱氢酶;(11):延胡索酸酶;(12):苹果酸脱氢酶;(13):乙酰辅酶A乙酰转移酶;(14):3-羟基丁酰辅酶A脱氢酶;(15):丁酰辅酶A脱氢酶;(16):3-羟基丁酰辅酶A脱水酶;(17):酰基辅酶A硫酯酶
Fig. 7 Distribution of DEPs in the central metabolic and caproic acid metabolic pathways “ +” indicates that the expression of the enzyme is up-regulated, “-” indicates that the expression of the enzyme is down-regulated. (1): Hexokinase.(2): ATP-dependent 6-phosphofructokinase.(3): Phosphoglycerate kinase.(4): Fructose diphosphate aldolase.(5): 2-phosphoglycerate dehydratase.(6): Pyruvate kinase.(7): Citrate synthase.(8): Isocitrate dehydrogenase.(9): α-ketoglutarate dehydrogenase complex.(10): Succinate dehydrogenase.(11): Fumarase.(12): Malate dehydrogenase.(13): Acetyl-CoA acetyltransferase.(14): 3-hydroxybutyryl-CoA dehydrogenase.(15): Butyryl-CoA dehydrogenase.(16): 3-hydroxybutyryl-CoA dehydratase.(17): Acyl-CoA thioesterase
| [1] |
游玲, 简晓平, 范方勇, 等. 宜宾浓香型白酒产区窖泥生态监测[J]. 生物技术通报, 2023, 39(7): 254-265.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1406 |
|
You L, Jian XP, Fan FY, et al. Ecological monitoring of pit mud in Yibin strong-fragrance baijiu-producing region[J]. Biotechnol Bull, 2023, 39(7): 254-265.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1406 |
|
| [2] | 孙宝国, 吴继红, 黄明泉, 等. 白酒风味化学研究进展[J]. 中国食品学报, 2015, 15(9): 1-8. |
| Sun BG, Wu JH, Huang MQ, et al. Recent advances of flavor chemistry in Chinese liquor spirits(baijiu)[J]. J Chin Inst Food Sci Technol, 2015, 15(9): 1-8. | |
| [3] |
赵辉, 敞颜, 等. 浓香型白酒窖泥中高产己酸兼性厌氧细菌的分离鉴定[J]. 食品科学, 2012, 33(5): 177-182.
doi: 10.7506/spkx1002-6630-201205038 |
| Zhao H, Chang Y, et al. Isolation and identification of facultative anaerobic strains with high yield of hexanoic acid from luzhou-flavor liquor pit mud[J]. Food Sci, 2012, 33(5): 177-182. | |
| [4] |
Yao F, Yi B, Shen CH, et al. Chemical analysis of the Chinese liquor Luzhoulaojiao by comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry[J]. Sci Rep, 2015, 5: 9553.
doi: 10.1038/srep09553 pmid: 25857434 |
| [5] | 陈茂彬, 殷想想, 郭志豪, 等. 酪丁酸梭菌共培养对速生梭菌己酸代谢的影响及机理探讨[J]. 食品科学, 2023, 44(10): 158-164. |
|
Chen MB, Yin XX, et al. Effect and mechanism of co-culture with Clostridium tyrobutyricum on caproic acid mechanism in Clostridium celerecrescens[J]. Food Sci, 2023, 44(10): 158-164.
doi: 10.1111/jfds.1979.44.issue-1 URL |
|
| [6] |
Kenealy WR, Cao Y, Weimer PJ. Production of caproic acid by cocultures of ruminal cellulolytic bacteria and Clostridium kluyveri grown on cellulose and ethanol[J]. Appl Microbiol Biotechnol, 1995, 44(3-4): 507-513.
doi: 10.1007/s002530050590 pmid: 8597554 |
| [7] |
Dong WJ, Yang YL, Liu C, et al. Caproic acid production from anaerobic fermentation of organic waste - Pathways and microbial perspective[J]. Renew Sustain Energy Rev, 2023, 175: 113181.
doi: 10.1016/j.rser.2023.113181 URL |
| [8] | Debergh P, Van Dael M. Production of caproic acid from acetate and ethanol through microbial chain elongation: a techno-economic assessment[J]. Bioresour Technol Rep, 2022, 18: 101055. |
| [9] |
Agler MT, Spirito CM, Usack JG, et al. Chain elongation with reactor microbiomes: upgrading dilute ethanol to medium-chain carboxylates[J]. Energy Environ Sci, 2012, 5(8): 8189-8192.
doi: 10.1039/c2ee22101b URL |
| [10] |
Nzeteu CO, Coelho F, et al. Development of an enhanced chain elon-gation process for caproic acid production from waste-derived lactic acid and butyric acid[J]. J Clean Prod, 2022, 338: 130655.
doi: 10.1016/j.jclepro.2022.130655 URL |
| [11] |
Yang Q, Guo ST, Lu Q, et al. Butyryl/Caproyl-CoA: acetate CoA-transferase: cloning, expression and characterization of the key enzyme involved in medium-chain fatty acid biosynthesis[J]. Biosci Rep, 2021, 41(8): BSR20211135.
doi: 10.1042/BSR20211135 URL |
| [12] |
Her J, Kim J. Rummeliibacillus suwonensis sp. nov., isolated from soil collected in a mountain area of South Korea[J]. J Microbiol, 2013, 51(2): 268-272.
doi: 10.1007/s12275-013-3126-5 URL |
| [13] |
Hutchinson-Bunch C, Sanford JA, Hansen JR, et al. Assessment of TMT labeling efficiency in large-scale quantitative proteomics: the critical effect of sample pH[J]. ACS Omega, 2021, 6(19): 12660-12666.
doi: 10.1021/acsomega.1c00776 pmid: 34056417 |
| [14] |
Liu CJ, Du YF, Zheng J, et al. Production of caproic acid by Rummeliibacillus suwonensis 3B-1 isolated from the pit mud of strong-flavor Baijiu[J]. J Biotechnol, 2022, 358: 33-40.
doi: 10.1016/j.jbiotec.2022.08.017 URL |
| [15] |
Wiśniewski JR, Zougman A, Nagaraj N, et al. Universal sample preparation method for proteome analysis[J]. Nat Methods, 2009, 6(5): 359-362.
doi: 10.1038/nmeth.1322 pmid: 19377485 |
| [16] |
Sinclair J, Timms JF. Quantitative profiling of serum samples using TMT protein labelling, fractionation and LC-MS/MS[J]. Methods, 2011, 54(4): 361-369.
doi: 10.1016/j.ymeth.2011.03.004 pmid: 21397697 |
| [17] | 郭长明, 袁橙, 武彩红, 等. 应用iTRAQ定量蛋白质组学技术筛选无乳链球菌鱼源株与人源株差异表达蛋白[J]. 水产学报, 2018, 42(3): 442-451. |
| Guo CM, Yuan C, Wu CH, et al. Quantitative proteomic analysis of differential proteins in Streptococcus agalactiae piscine strain and human strain using iTRAQ[J]. J Fish China, 2018, 42(3): 442-451. | |
| [18] |
Rojas-Pirela M, Andrade-Alviárez D, et al. Phosphoglycerate kinase: structural aspects and functions, with special emphasis on the enzy-me from Kinetoplastea[J]. Open Biol, 2020, 10(11): 200302.
doi: 10.1098/rsob.200302 URL |
| [19] |
Yang LY, Wang ZX, Zhang AQ, et al. Reduction of the canonical function of a glycolytic enzyme enolase triggers immune responses that further affect metabolism and growth in Arabidopsis[J]. Plant Cell, 2022, 34(5): 1745-1767.
doi: 10.1093/plcell/koab283 URL |
| [20] |
Moxley WC, Eiteman MA. Pyruvate production by Escherichia coli by use of pyruvate dehydrogenase variants[J]. Appl Environ Microbiol, 2021, 87(13): e0048721.
doi: 10.1128/AEM.00487-21 URL |
| [21] |
Starling S. Pyruvate kinase regulates insulin secretion[J]. Nat Rev Endocrinol, 2021, 17(1): 3.
doi: 10.1038/s41574-020-00447-0 pmid: 33208920 |
| [22] |
Jacob H, Geng H, Shetty D, et al. Distinct interaction mechanism of RNA polymerase and ResD at proximal and distal subsites for transcription activation of nitrite reductase in Bacillus subtilis[J]. J Bacteriol, 2022, 204(2): e0043221.
doi: 10.1128/jb.00432-21 URL |
| [23] |
Nakano MM, Zuber P, Glaser P, et al. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis[J]. J Bacteriol, 1996, 178(13): 3796-3802.
doi: 10.1128/jb.178.13.3796-3802.1996 pmid: 8682783 |
| [24] |
Michalska K, Jedrzejczak R, Wower J, et al. Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation[J]. Nucleic Acids Res, 2021, 49(9): 5351-5368.
doi: 10.1093/nar/gkab272 pmid: 33885823 |
| [25] |
Cong Q, Anishchenko I, Ovchinnikov S, et al. Protein interaction networks revealed by proteome coevolution[J]. Science, 2019, 365(6449): 185-189.
doi: 10.1126/science.aaw6718 pmid: 31296772 |
| [26] |
Sepúlveda-Cisternas I, Lozano Aguirre L, Fuentes Flores A, et al. Transcriptomics reveals a cross-modulatory effect between riboflavin and iron and outlines responses to riboflavin biosynthesis and uptake in Vibrio cholerae[J]. Sci Rep, 2018, 8(1): 3149.
doi: 10.1038/s41598-018-21302-3 pmid: 29453341 |
| [27] |
Yu S, Meng SM, Xiang MX, et al. Phosphoenolpyruvate carboxykinase in cell metabolism: roles and mechanisms beyond gluconeogenesis[J]. Mol Metab, 2021, 53: 101257.
doi: 10.1016/j.molmet.2021.101257 URL |
| [28] |
Britt EC, Lika J, Giese MA, et al. Switching to the cyclic pentose phosphate pathway powers the oxidative burst in activated neutrophils[J]. Nat Metab, 2022, 4(3): 389-403.
doi: 10.1038/s42255-022-00550-8 pmid: 35347316 |
| [29] |
Carvajal-Arroyo JM, Candry P, Andersen SJ, et al. Granular fermentation enables high rate caproic acid production from solid-free thin stillage[J]. Green Chem, 2019, 21(6): 1330-1339.
doi: 10.1039/c8gc03648a |
| [30] |
Chwialkowska J, Duber A, Zagrodnik R, et al. Caproic acid production from acid whey via open culture fermentation - Evaluation of the role of electron donors and downstream processing[J]. Bioresour Technol, 2019, 279: 74-83.
doi: 10.1016/j.biortech.2019.01.086 URL |
| [31] |
徐友强, 孙宝国, 蒋玥凤, 等. 基于比较基因组学的Clostrid-ium kluyveri己酸代谢途径关键酶生物信息学分析[J]. 食品科学, 2019, 40(4): 122-129.
doi: 10.7506/spkx1002-6630-20180413-183 |
| Xu YQ, Sun BG, Jiang YF, et al. Bioinformatics analysis of the key enzymes of the hexanoic acid metabolic pathway in Clostridium kluyveri based on comparative genomics[J]. Food Sci, 2019, 40(4): 122-129. |
| [1] | 杨伟杰, 杨周林, 朱浩东, 魏煜, 刘君, 刘训. 地衣素合成酶关键模块 LchAD 蛋白的性质和功能研究[J]. 生物技术通报, 2024, 40(3): 322-332. |
| [2] | 龚丽丽, 余花, 杨杰, 陈天池, 赵双滢, 吴月燕. 葡萄CYP707A基因家族的鉴定及对果实成熟的功能验证[J]. 生物技术通报, 2024, 40(2): 160-171. |
| [3] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [4] | 李敬蕊, 王育博, 解紫薇, 李畅, 吴晓蕾, 宫彬彬, 高洪波. 甜瓜PIN基因家族的鉴定及高温胁迫表达分析[J]. 生物技术通报, 2023, 39(5): 192-204. |
| [5] | 郭三保, 宋美玲, 李灵心, 尧子钊, 桂明明, 黄胜和. 斑地锦查尔酮合酶基因及启动子的克隆与分析[J]. 生物技术通报, 2023, 39(4): 148-156. |
| [6] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [7] | 杨岚, 张晨曦, 樊学伟, 王阳光, 王春秀, 李文婷. 鸡 BMP15 基因克隆、表达模式及启动子活性分析[J]. 生物技术通报, 2023, 39(4): 304-312. |
| [8] | 陈强, 邹明康, 宋家敏, 张冲, 吴隆坤. 甜瓜LBD基因家族的鉴定和果实发育进程中的表达分析[J]. 生物技术通报, 2023, 39(3): 176-183. |
| [9] | 平怀磊, 郭雪, 余潇, 宋静, 杜春, 王娟, 张怀璧. 滇牡丹PdANS的克隆、表达及与花青素含量的相关性[J]. 生物技术通报, 2023, 39(3): 206-217. |
| [10] | 邢媛, 宋健, 李俊怡, 郑婷婷, 刘思辰, 乔治军. 谷子AP基因家族鉴定及其对非生物胁迫的响应分析[J]. 生物技术通报, 2023, 39(11): 238-251. |
| [11] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [12] | 杨敏, 龙雨青, 曾娟, 曾梅, 周新茹, 王玲, 付学森, 周日宝, 刘湘丹. 灰毡毛忍冬UGTPg17、UGTPg36基因克隆及功能研究[J]. 生物技术通报, 2023, 39(10): 256-267. |
| [13] | 郭志浩, 金泽鑫, 刘琦, 高利. 小麦矮腥黑粉菌效应蛋白g11335的生物信息学分析、亚细胞定位及毒性验证[J]. 生物技术通报, 2022, 38(8): 110-117. |
| [14] | 于秋琳, 马婧怡, 赵盼, 孙鹏芳, 何玉美, 刘世彪, 郭惠红. 绞股蓝GpMIR156a和GpMIR166b的克隆与功能分析[J]. 生物技术通报, 2022, 38(7): 186-193. |
| [15] | 陈佳敏, 刘永杰, 马锦绣, 李丹, 公杰, 赵昌平, 耿洪伟, 高世庆. 小麦组蛋白甲基化酶在杂交种中干旱胁迫表达模式分析[J]. 生物技术通报, 2022, 38(7): 51-61. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||