








生物技术通报 ›› 2024, Vol. 40 ›› Issue (7): 19-27.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0109
杨嘉泓1( ), 李婧怡1, 吴佳昊2, 黄幼梅1, 刘艳芬1, 秦源1(
), 李婧怡1, 吴佳昊2, 黄幼梅1, 刘艳芬1, 秦源1( ), 蔡汉阳1(
), 蔡汉阳1( )
)
收稿日期:2024-01-30
出版日期:2024-07-26
发布日期:2024-07-30
通讯作者:
秦源,女,博士,教授,研究方向:植物生殖发育;E-mail: yuanqin@fafu.edu.cn;作者简介:杨嘉泓,女,硕士研究生,研究方向:植物生殖发育;E-mail: yangjiahong1224@163.com
基金资助:
YANG Jia-hong1( ), LI Jing-yi1, WU Jia-hao2, HUANG You-mei1, LIU Yan-fen1, QIN Yuan1(
), LI Jing-yi1, WU Jia-hao2, HUANG You-mei1, LIU Yan-fen1, QIN Yuan1( ), CAI Han-yang1(
), CAI Han-yang1( )
)
Received:2024-01-30
Published:2024-07-26
Online:2024-07-30
摘要:
雌配子体发育是开花植物有性生殖的重要前提,包括大孢子发生和雌配子体发生两个关键事件,该过程决定植物能否正常受精作用并完成世代繁衍。生长素是植物中广泛存在的一类植物激素,生长素在胚珠中的极性分布保证了胚珠的正常发育,但由于生长素信号途径十分复杂,且雌性生殖细胞位于组织内部,生长素信号途径在雌性生殖系建成的调控网络和分子机制仍不明晰。本文综述了生长素的合成、动态平衡和信号转导以及生长素调控大孢子发生和雌配子体发生相关研究,旨在对生长素激素信号途径调控雌性生殖系建成提供参考。
杨嘉泓, 李婧怡, 吴佳昊, 黄幼梅, 刘艳芬, 秦源, 蔡汉阳. 生长素信号途径参与调控拟南芥雌配子体发育研究进展[J]. 生物技术通报, 2024, 40(7): 19-27.
YANG Jia-hong, LI Jing-yi, WU Jia-hao, HUANG You-mei, LIU Yan-fen, QIN Yuan, CAI Han-yang. Research Progress in the Auxin Signaling Pathway Involved in the Regulation of Female Gametophyte Development in Arabidopsis[J]. Biotechnology Bulletin, 2024, 40(7): 19-27.
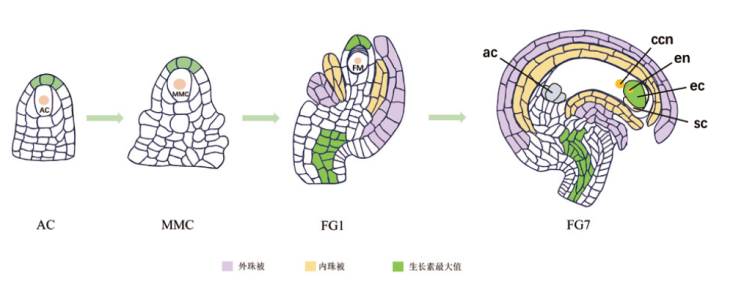
图2 拟南芥胚珠大孢子发生时期和雌配子体发生时期的生长素极性分布 AC:孢原细胞;MMC:大孢子母细胞;FM:功能大孢子;ac:反足细胞;ccn:中央极核;en:卵细胞核;ec:卵细胞;sc:助细胞
Fig. 2 Polarity distribution of auxin during the period of ovule megasporogenesis and female gametophyte genesis in Arabidopsis thaliana AC: Archesporial cell; MMC: megaspore mother cell; FM: functional megaspore; ac: antipodal cell; ccn: central polar nucellus; en: egg cell nucleus; ec: egg cell; sc: synergid cell
| [1] | Yang WC, Shi DQ, Chen YH. Female gametophyte development in flowering plants[J]. Annu Rev Plant Biol, 2010, 61: 89-108. |
| [2] |
Böwer F, Schnittger A. How to switch from mitosis to meiosis: regulation of germline entry in plants[J]. Annu Rev Genet, 2021, 55: 427-452.
doi: 10.1146/annurev-genet-112618-043553 pmid: 34530640 |
| [3] |
Pinto SC, Mendes MA, Coimbra S, et al. Revisiting the female germline and its expanding toolbox[J]. Trends Plant Sci, 2019, 24(5): 455-467.
doi: S1360-1385(19)30030-5 pmid: 30850278 |
| [4] |
Zhao YD. Auxin biosynthesis and its role in plant development[J]. Annu Rev Plant Biol, 2010, 61: 49-64.
doi: 10.1146/annurev-arplant-042809-112308 pmid: 20192736 |
| [5] | Luo P, Di DW. Precise regulation of the TAA1/TAR-YUCCA auxin biosynthesis pathway in plants[J]. Int J Mol Sci, 2023, 24(10): 8514. |
| [6] |
Tao Y, Ferrer JL, Ljung K, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants[J]. Cell, 2008, 133(1):164-176.
doi: 10.1016/j.cell.2008.01.049 pmid: 18394996 |
| [7] |
Stepanova AN, Robertson-Hoyt J, Yun J, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development[J]. Cell, 2008, 133(1): 177-191.
doi: 10.1016/j.cell.2008.01.047 pmid: 18394997 |
| [8] | Di DW, Wu L, Zhang L, et al. Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin[J]. Sci Rep, 2016, 6: 36866. |
| [9] | Cao X, Yang HL, Shang CQ, et al. The roles of auxin biosynthesis YUCCA gene family in plants[J]. Int J Mol Sci, 2019, 20(24): 6343. |
| [10] |
Zhao Y, Christensen SK, Fankhauser C, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis[J]. Science, 2001, 291(5502): 306-309.
doi: 10.1126/science.291.5502.306 pmid: 11209081 |
| [11] | Panoli A, Martin MV, Alandete-Saez M, et al. Auxin import and local auxin biosynthesis are required for mitotic divisions, cell expansion and cell specification during female gametophyte development in Arabidopsis thaliana[J]. PLoS One, 2015, 10(5): e0126164. |
| [12] |
Staswick PE, Serban B, Rowe M, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid[J]. Plant Cell, 2005, 17(2): 616-627.
doi: 10.1105/tpc.104.026690 pmid: 15659623 |
| [13] |
Ludwig-Müller J. Auxin conjugates: their role for plant development and in the evolution of land plants[J]. J Exp Bot, 2011, 62(6): 1757-1773.
doi: 10.1093/jxb/erq412 pmid: 21307383 |
| [14] | Zhang SW, Li CH, Cao J, et al. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation[J]. Plant Physiol, 2009, 151(4): 1889-1901. |
| [15] |
Wang Q, Qin GC, Cao M, et al. A phosphorylation-based switch controls TAA1-mediated auxin biosynthesis in plants[J]. Nat Commun, 2020, 11(1): 679.
doi: 10.1038/s41467-020-14395-w pmid: 32015349 |
| [16] |
Robert HS, Friml J. Auxin and other signals on the move in plants[J]. Nat Chem Biol, 2009, 5(5): 325-332.
doi: 10.1038/nchembio.170 pmid: 19377459 |
| [17] | Mravec J, Skůpa P, Bailly A, et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter[J]. Nature, 2009, 459(7250): 1136-1140. |
| [18] |
Mravec J, Kubes M, Bielach A, et al. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development[J]. Development, 2008, 135(20): 3345-3354.
doi: 10.1242/dev.021071 pmid: 18787070 |
| [19] |
Petrásek J, Mravec J, Bouchard R, et al. PIN proteins perform a rate-limiting function in cellular auxin efflux[J]. Science, 2006, 312(5775): 914-918.
doi: 10.1126/science.1123542 pmid: 16601150 |
| [20] | Yang ZS, Xia J, Hong JJ, et al. Structural insights into auxin recognition and efflux by Arabidopsis PIN1[J]. Nature, 2022, 609(7927): 611-615. |
| [21] | Su NN, Zhu AQ, Tao X, et al. Structures and mechanisms of the Arabidopsis auxin transporter PIN3[J]. Nature, 2022, 609(7927): 616-621. |
| [22] | Bainbridge K, Guyomarc'h S, Bayer E, et al. Auxin influx carriers stabilize phyllotactic patterning[J]. Genes Dev, 2008, 22(6): 810-823. |
| [23] |
Yang YD, Hammes UZ, Taylor CG, et al. High-affinity auxin transport by the AUX1 influx carrier protein[J]. Curr Biol, 2006, 16(11): 1123-1127.
doi: 10.1016/j.cub.2006.04.029 pmid: 16677815 |
| [24] |
Titapiwatanakun B, Murphy AS. Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition[J]. J Exp Bot, 2009, 60(4): 1093-1107.
doi: 10.1093/jxb/ern240 pmid: 18824505 |
| [25] | Ceccato L, Masiero S, Sinha Roy D, et al. Maternal control of PIN1 is required for female gametophyte development in Arabidopsis[J]. PLoS One, 2013, 8(6): e66148. |
| [26] | Wu WT, Liu YX, Wang YQ, et al. Evolution analysis of the aux/IAA gene family in plants shows dual origins and variable nuclear localization signals[J]. Int J Mol Sci, 2017, 18(10): 2107. |
| [27] |
Ito J, Fukaki H, Onoda M, et al. Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription[J]. Proc Natl Acad Sci USA, 2016, 113(23): 6562-6567.
doi: 10.1073/pnas.1600739113 pmid: 27217573 |
| [28] |
Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis[J]. Science, 2008, 319(5868): 1384-1386.
doi: 10.1126/science.1151461 pmid: 18258861 |
| [29] | Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor[J]. Nature, 2005, 435(7041): 446-451. |
| [30] |
Winkler M, Niemeyer M, Hellmuth A, et al. Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction[J]. Nat Commun, 2017, 8: 15706.
doi: 10.1038/ncomms15706 pmid: 28589936 |
| [31] | Dezfulian MH, Jalili E, Roberto DKA, et al. Oligomerization of SCFTIR1 is essential for aux/IAA degradation and auxin signaling in Arabidopsis[J]. PLoS Genet, 2016, 12(9): e1006301. |
| [32] | Luo J, Zhou JJ, Zhang JZ. Aux/IAA gene family in plants: molecular structure, regulation, and function[J]. Int J Mol Sci, 2018, 19(1): 259. |
| [33] |
Han M, Park Y, Kim I, et al. Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17[J]. Proc Natl Acad Sci USA, 2014, 111(52): 18613-18618.
doi: 10.1073/pnas.1419525112 pmid: 25512488 |
| [34] | Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors[J]. Plant Mol Biol, 2002, 49(3/4): 373-385. |
| [35] |
Guilfoyle TJ, Hagen G. Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions[J]. Plant Sci, 2012, 190: 82-88.
doi: 10.1016/j.plantsci.2012.04.003 pmid: 22608522 |
| [36] |
Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins[J]. Proc Natl Acad Sci USA, 1997, 94(22): 11786-11791.
doi: 10.1073/pnas.94.22.11786 pmid: 9342315 |
| [37] | Tan X, Calderon-Villalobos LIA, Sharon M, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase[J]. Nature, 2007, 446(7136): 640-645. |
| [38] | Salehin M, Bagchi R, Estelle M. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development[J]. Plant Cell, 2015, 27(1): 9-19. |
| [39] | Li SB, Xie ZZ, Hu CG, et al. A review of auxin response factors(ARFs)in plants[J]. Front Plant Sci, 2016, 7: 47. |
| [40] |
Guilfoyle TJ, Hagen G. Auxin response factors[J]. Curr Opin Plant Biol, 2007, 10(5): 453-460.
doi: 10.1016/j.pbi.2007.08.014 pmid: 17900969 |
| [41] |
Yu ZP, Zhang F, Friml J, et al. Auxin signaling: research advances over the past 30 years[J]. J Integr Plant Biol, 2022, 64(2): 371-392.
doi: 10.1111/jipb.13225 |
| [42] |
Chen HH, Li LX, Zou MX, et al. Distinct functions of TIR1 and AFB1 receptors in auxin signaling[J]. Mol Plant, 2023, 16(7): 1117-1119.
doi: 10.1016/j.molp.2023.06.007 pmid: 37393433 |
| [43] | Qi LL, Kwiatkowski M, Chen HH, et al. Adenylate cyclase activity of TIR1/AFB auxin receptors in plants[J]. Nature, 2022, 611(7934): 133-138. |
| [44] | Lin WW, Zhou X, Tang WX, et al. TMK-based cell-surface auxin signalling activates cell-wall acidification[J]. Nature, 2021, 599(7884): 278-282. |
| [45] | Friml J, Gallei M, Gelová Z, et al. ABP1-TMK auxin perception for global phosphorylation and auxin canalization[J]. Nature, 2022, 609(7927): 575-581. |
| [46] |
Yu YQ, Tang WX, Lin WW, et al. ABLs and TMKs are co-receptors for extracellular auxin[J]. Cell, 2023, 186(25): 5457-5471.e17.
doi: 10.1016/j.cell.2023.10.017 pmid: 37979582 |
| [47] | Wang LL, Liu YH, Aslam M, et al. The Glycine-rich domain protein GRDP2 regulates ovule development via the auxin pathway in Arabidopsis[J]. Front Plant Sci, 2021, 12: 698487. |
| [48] | Cai HY, Liu LP, Ma SZ, et al. Insights into the role of phytohormones in plant female germline cell specification[J]. Curr Opin Plant Biol, 2023, 75: 102439. |
| [49] | Sun Y, Wang X, Pan L, et al. Plant egg cell fate determination depends on its exact position in female gametophyte[J]. Proc Natl Acad Sci USA, 2021, 118(8): e2017488118. |
| [50] | Jiang YT, Zheng JX, Li RH, et al. Tonoplast proton pumps regulate nuclear spacing of female gametophytes via mediating polar auxin transport in Arabidopsis[J]. Front Plant Sci, 2022, 13: 1006735. |
| [51] | Rojek J, Tucker MR, Pinto SC, et al. Rab-dependent vesicular traffic affects female gametophyte development in Arabidopsis[J]. J Exp Bot, 2021, 72(2): 320-340. |
| [52] |
Huang J, Zhao L, Malik S, et al. Specification of female germline by microRNA orchestrated auxin signaling in Arabidopsis[J]. Nat Commun, 2022, 13(1): 6960.
doi: 10.1038/s41467-022-34723-6 pmid: 36379956 |
| [53] | Abas L, Kolb M, Stadlmann J, et al. Naphthylphthalamic acid associates with and inhibits PIN auxin transporters[J]. Proc Natl Acad Sci USA, 2021, 118(1): e2020857118. |
| [54] |
Su ZX, Zhao LH, Zhao YY, et al. The THO complex non-cell-autonomously represses female germline specification through the TAS3-ARF3 module[J]. Curr Biol, 2017, 27(11): 1597-1609.e2.
doi: S0960-9822(17)30557-2 pmid: 28552357 |
| [55] | Su ZX, Wang NN, Hou ZM, et al. Regulation of female germline specification via small RNA mobility in Arabidopsis[J]. Plant Cell, 2020, 32(9): 2842-2854. |
| [56] | Liu ZN, Miao LM, Huo RX, et al. ARF2-ARF4 and ARF5 are essential for female and male gametophyte development in Arabidopsis[J]. Plant Cell Physiol, 2018, 59(1): 179-189. |
| [57] |
Kieffer M, Neve J, Kepinski S. Defining auxin response contexts in plant development[J]. Curr Opin Plant Biol, 2010, 13(1): 12-20.
doi: 10.1016/j.pbi.2009.10.006 pmid: 19942473 |
| [58] | Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis[J]. Nature, 2008, 453(7198): 1094-1097. |
| [59] |
Vert G, Walcher CL, Chory J, et al. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2[J]. Proc Natl Acad Sci USA, 2008, 105(28): 9829-9834.
doi: 10.1073/pnas.0803996105 pmid: 18599455 |
| [60] | Qadir M, Wang XF, Shah SRU, et al. Molecular network for regulation of ovule number in plants[J]. Int J Mol Sci, 2021, 22(23): 12965. |
| [61] |
Lora J, Yang XJ, Tucker MR. Establishing a framework for female germline initiation in the plant ovule[J]. J Exp Bot, 2019, 70(11): 2937-2949.
doi: 10.1093/jxb/erz212 pmid: 31063548 |
| [62] | Bencivenga S, Simonini S, Benková E, et al. The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis[J]. Plant Cell, 2012, 24(7): 2886-2897. |
| [63] |
Cucinotta M, Di Marzo M, Guazzotti A, et al. Gynoecium size and ovule number are interconnected traits that impact seed yield[J]. J Exp Bot, 2020, 71(9): 2479-2489.
doi: 10.1093/jxb/eraa050 pmid: 32067041 |
| [64] |
Cucinotta M, Manrique S, Guazzotti A, et al. Cytokinin response factors integrate auxin and cytokinin pathways for female reproductive organ development[J]. Development, 2016, 143(23): 4419-4424.
pmid: 27737904 |
| [65] | Wu MF, Tian Q, Reed JW. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction[J]. Development, 2006, 133(21): 4211-4218. |
| [1] | 谢彦杰. 和而不同:根毛发育过程中生长素与氧化还原信号[J]. 生物技术通报, 2024, 40(6): 1-4. |
| [2] | 王贤, 彭亚坤, 陈猛, 孔梦娟, 谭树堂. 植物向重力反应中PIN-FORMED介导的生长素极性运输调控[J]. 生物技术通报, 2024, 40(3): 25-40. |
| [3] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [4] | 杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142. |
| [5] | 胡琪, 侯玉翔, 李璿, 李梅兰. 普通白菜CYP79B2同源基因的克隆与表达[J]. 生物技术通报, 2022, 38(12): 168-174. |
| [6] | 钱静洁, 林苏梦, 张冬平, 高勇. 光敏色素互作因子参与生长素调控的植物生长发育[J]. 生物技术通报, 2022, 38(10): 29-33. |
| [7] | 唐嘉城, 梁毅珉, 马葭思, 彭桂香, 谭志远. 百香果内生细菌多样性及促生长特性[J]. 生物技术通报, 2022, 38(1): 86-97. |
| [8] | 周希萌, 付春, 马长乐, 王兴军, 赵传志. 作物分枝的分子调控研究进展[J]. 生物技术通报, 2021, 37(3): 107-114. |
| [9] | 冯寒骞, 李超. 生长素信号转导研究进展[J]. 生物技术通报, 2018, 34(7): 24-30. |
| [10] | 窦悦, 刘美彤, 卢安娜, 吴佳洁, 王群青, 胥倩. 中介体亚基MED25调控植物激素信号转导的研究进展[J]. 生物技术通报, 2018, 34(7): 40-47. |
| [11] | 侯鹏飞,贾振华,宋水山,. 生长素和细胞分裂素调控植物根和微生物互作的研究进展[J]. 生物技术通报, 2017, 33(7): 1-6. |
| [12] | 葛伟娜, 李超, 张家琦, 张岚, 王振怡. 植物天冬氨酸蛋白酶的研究进展[J]. 生物技术通报, 2016, 32(1): 8-14. |
| [13] | 孙帆, 罗朝兵, 周燕妮, 张凌云. 青杄生长素抑制蛋白基因PwARP-1的克隆及表达分析[J]. 生物技术通报, 2014, 30(4): 64-70. |
| [14] | 李静;崔继哲;弭晓菊;. 生长素与植物逆境胁迫关系的研究进展[J]. , 2012, 0(06): 13-17. |
| [15] | 任怡怡;戴绍军;刘炜;. 生长素的运输及其在信号转导及植物发育中的作用[J]. , 2012, 0(03): 9-16. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||