








生物技术通报 ›› 2024, Vol. 40 ›› Issue (8): 199-211.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0139
王茜( ), 周家燕, 王倩, 邓玉萍, 张敏慧, 陈静, 杨军, 邹建(
), 周家燕, 王倩, 邓玉萍, 张敏慧, 陈静, 杨军, 邹建( )
)
收稿日期:2024-02-04
出版日期:2024-08-26
发布日期:2024-06-26
通讯作者:
邹建,男,博士,副教授,研究方向:植物遗传与发育;E-mail: zoujian@cwnu.edu.cn作者简介:王茜,女,硕士研究生,研究方向:植物遗传与发育;E-mail: 1347404972@qq.com
基金资助:
WANG Qian( ), ZHOU Jia-yan, WANG Qian, DENG Yu-ping, ZHANG Min-hui, CHEN Jing, YANG Jun, ZOU Jian(
), ZHOU Jia-yan, WANG Qian, DENG Yu-ping, ZHANG Min-hui, CHEN Jing, YANG Jun, ZOU Jian( )
)
Received:2024-02-04
Published:2024-08-26
Online:2024-06-26
摘要:
【目的】鉴定向日葵中花发育相关的HaYABBY基因,有助于探究其在向日葵花发育过程中的生物学功能和调控机制,为向日葵品种选育提供重要的分子线索。【方法】基于向日葵基因组和转录组数据,使用生物信息学方法,分析向日葵YABBY基因家族成员的染色体定位、系统进化关系、基因结构、保守基序、蛋白结构及理化性质、启动子顺式作用元件和组织表达模式等,并通过RT-qPCR分析HaYABBY基因在WT向日葵和花分生组织决定和花器官分化缺陷突变体cb1中st2-st8时期的表达差异。【结果】在向日葵基因组中鉴定到12个YABBY基因家族成员,分属于4个亚家族,分布在8条染色体上,并且同一亚家族中成员在基因结构和保守基序组成上高度相似。其中8个基因HaYABBY1a、HaYABBY1c、HaYABBY3、HaYABBY4b、HaYABBY5a、HaYABBY5b、HaCRABS CLAWa、HaCRABS CLAWb在向日葵花发育时期特异性高表达。在这8个基因中,HaYABBY3、HaYABBY5a和HaYABBY5d在突变体cb1花发育后期表达上调,且与WT花发育早期水平相当;而HaYABBY4b和HaCRABS CLAWa在cb1中的表达被强烈抑制。顺式作用元件分析显示,HaYABBY均含有多个启动子顺式作用元件,部分元件仅存在于特定基因中。其中,胚乳表达元件存在于HaCRABS CLAWa和HaYABBY4a中;生长素响应元件存在于HaCRABS CLAWa、HaCRABS CLAWb、HaYABBY4a、HaYABBY4b和HaYABBY5c中;分生组织表达元件存在于HaYABBY1a、HaYABBY1c、HaYABBY4b、HaYABBY5a和HaYABBY5d中;赤霉素响应元件存在于HaCRABS CLAWa、HaYABBY5b、HaYABBY5c和HaYABBY5d中;种子特异性调控元件存在于HaYABBY5a、HaYABBY5c和HaYABBY5d中。【结论】HaYABBY3、HaYABBY4b、HaYABBY5a、HaYABBY5d和HaCRABS CLAWa等5个HaYABBY基因在花分生组织的转变和花器官发生过程中可能发挥重要作用,它们在花分生组织转变和花器官发生调控中可能受到生长素响应因子ARF、赤霉素响应因子MYB以及MADS-box等关键因子调节。
王茜, 周家燕, 王倩, 邓玉萍, 张敏慧, 陈静, 杨军, 邹建. 向日葵YABBY基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(8): 199-211.
WANG Qian, ZHOU Jia-yan, WANG Qian, DENG Yu-ping, ZHANG Min-hui, CHEN Jing, YANG Jun, ZOU Jian. Identification and Expression Analysis of the YABBY Gene Family in Sunflower[J]. Biotechnology Bulletin, 2024, 40(8): 199-211.
| 基因 Gene | 基因编号 Gene ID | 染色体分布Chromosome distribution | 氨基酸数目 Number of amino acids | 等电点 pI | 分子量 Molecular weight/Da | 亚细胞定位 Subcellular location | ||
|---|---|---|---|---|---|---|---|---|
| 编号No. | 开始位置Starting | 结束位置Ending | ||||||
| HaYABBY 1a | HanXRQr2_Chr06g0245541 | 6 | 12 754 239 | 12 758 093 | 219 | 7.21 | 24 420.73 | 细胞核 |
| HaYABBY 1b | HanXRQr2_Chr13g0613121 | 13 | 161 766 031 | 161 771 918 | 216 | 6.72 | 24 136.35 | 细胞核 |
| HaYABBY 1c | HanXRQr2_Chr04g0185291 | 4 | 195 371 365 | 195 376 174 | 219 | 7.21 | 24 437.79 | 细胞核 |
| HaCRABS CLAWa | HanXRQr2_Chr12g0549931 | 1 | 81 233 913 | 81 243 999 | 173 | 9.4 | 19 366.25 | 细胞核 |
| HaCRABS CLAWb | HanXRQr2_Chr15g0688211 | 15 | 39 093 254 | 39 095 798 | 180 | 9.28 | 19 765.53 | 细胞核 |
| HaYABBY 3 | HanXRQr2_Chr13g0577751 | 13 | 57 727 478 | 57 739 518 | 225 | 9.16 | 24 724.41 | 细胞核 |
| HaYABBY 4a | HanXRQr2_Chr17g0787111 | 17 | 15 387 218 | 15 388 714 | 197 | 4.98 | 21 878.58 | 细胞核 |
| HaYABBY 4b | HanXRQr2_Chr06g0253091 | 6 | 30 699 177 | 3 700 575 | 236 | 6.4 | 26 603.38 | 细胞核 |
| HaYABBY 4c | HanXRQr2_Chr03g0102331 | 3 | 78 930 712 | 78 931 672 | 99 | 7.82 | 10 936.49 | 细胞核 |
| HaYABBY 5a | HanXRQr2_Chr17g0801501 | 17 | 53 868 011 | 53 872 537 | 227 | 8.55 | 25 164.39 | 细胞核 |
| HaYABBY 5b | HanXRQr2_Chr10g0462601 | 10 | 175 418 675 | 175 424 195 | 200 | 9.74 | 22 555.59 | 细胞核 |
| HaYABBY 5c | HanXRQr2_Chr12g0523971 | 12 | 5 127 939 | 5 135 711 | 186 | 9.67 | 21 053.74 | 细胞核 |
| HaYABBY 5d | HanXRQr2_Chr06g0238981 | 6 | 1 174 948 | 1 181 807 | 229 | 8.5 | 25 139.60 | 细胞核 |
表1 向日葵YABBY基因家族成员信息
Table 1 Information of YABBY gene family members in sunflower
| 基因 Gene | 基因编号 Gene ID | 染色体分布Chromosome distribution | 氨基酸数目 Number of amino acids | 等电点 pI | 分子量 Molecular weight/Da | 亚细胞定位 Subcellular location | ||
|---|---|---|---|---|---|---|---|---|
| 编号No. | 开始位置Starting | 结束位置Ending | ||||||
| HaYABBY 1a | HanXRQr2_Chr06g0245541 | 6 | 12 754 239 | 12 758 093 | 219 | 7.21 | 24 420.73 | 细胞核 |
| HaYABBY 1b | HanXRQr2_Chr13g0613121 | 13 | 161 766 031 | 161 771 918 | 216 | 6.72 | 24 136.35 | 细胞核 |
| HaYABBY 1c | HanXRQr2_Chr04g0185291 | 4 | 195 371 365 | 195 376 174 | 219 | 7.21 | 24 437.79 | 细胞核 |
| HaCRABS CLAWa | HanXRQr2_Chr12g0549931 | 1 | 81 233 913 | 81 243 999 | 173 | 9.4 | 19 366.25 | 细胞核 |
| HaCRABS CLAWb | HanXRQr2_Chr15g0688211 | 15 | 39 093 254 | 39 095 798 | 180 | 9.28 | 19 765.53 | 细胞核 |
| HaYABBY 3 | HanXRQr2_Chr13g0577751 | 13 | 57 727 478 | 57 739 518 | 225 | 9.16 | 24 724.41 | 细胞核 |
| HaYABBY 4a | HanXRQr2_Chr17g0787111 | 17 | 15 387 218 | 15 388 714 | 197 | 4.98 | 21 878.58 | 细胞核 |
| HaYABBY 4b | HanXRQr2_Chr06g0253091 | 6 | 30 699 177 | 3 700 575 | 236 | 6.4 | 26 603.38 | 细胞核 |
| HaYABBY 4c | HanXRQr2_Chr03g0102331 | 3 | 78 930 712 | 78 931 672 | 99 | 7.82 | 10 936.49 | 细胞核 |
| HaYABBY 5a | HanXRQr2_Chr17g0801501 | 17 | 53 868 011 | 53 872 537 | 227 | 8.55 | 25 164.39 | 细胞核 |
| HaYABBY 5b | HanXRQr2_Chr10g0462601 | 10 | 175 418 675 | 175 424 195 | 200 | 9.74 | 22 555.59 | 细胞核 |
| HaYABBY 5c | HanXRQr2_Chr12g0523971 | 12 | 5 127 939 | 5 135 711 | 186 | 9.67 | 21 053.74 | 细胞核 |
| HaYABBY 5d | HanXRQr2_Chr06g0238981 | 6 | 1 174 948 | 1 181 807 | 229 | 8.5 | 25 139.60 | 细胞核 |
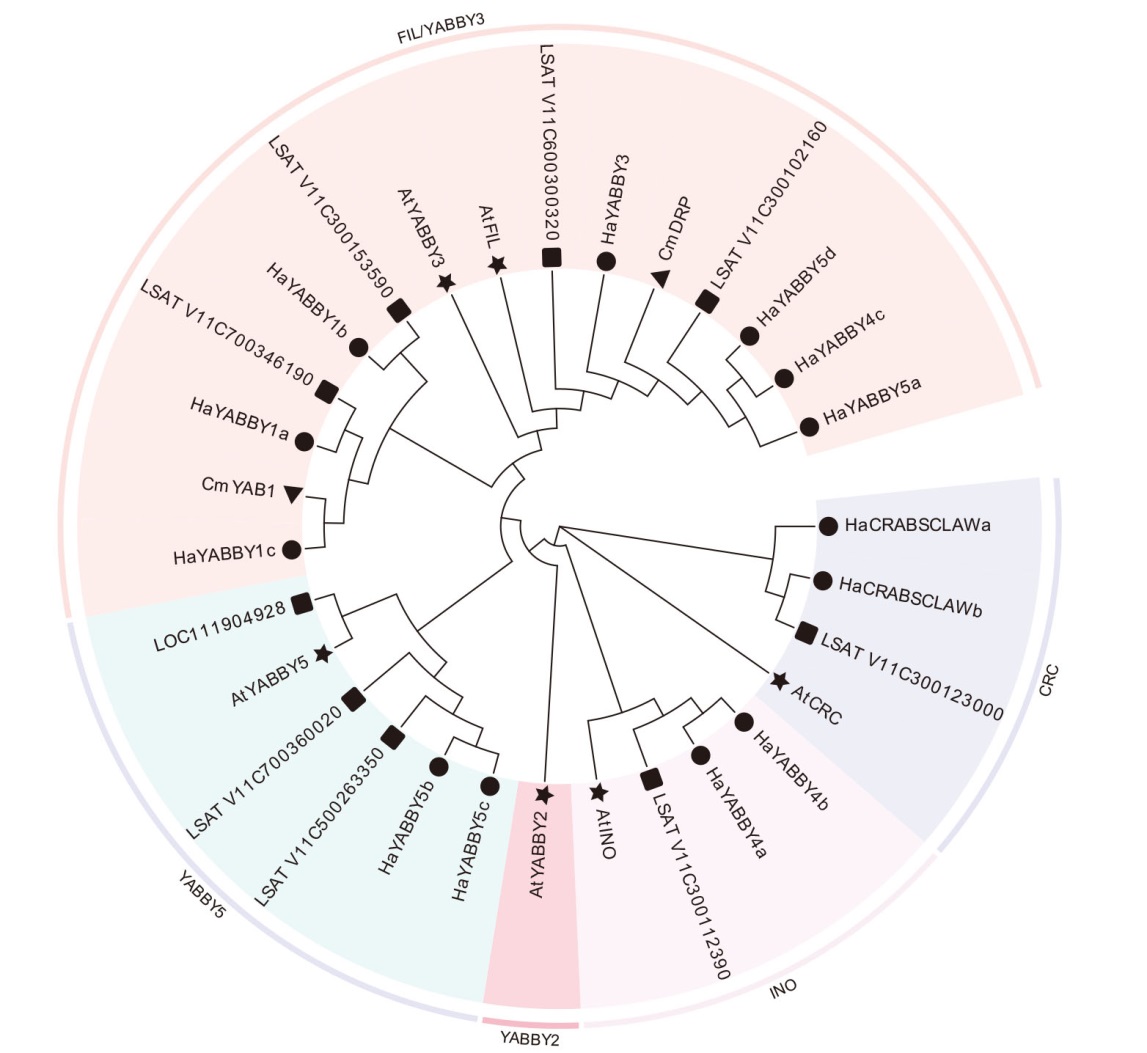
图2 向日葵YABBY基因家族成员的系统发育树分析 ★代表拟南芥;●代表向日葵;▲代表菊花;■代表莴苣
Fig. 2 Phylogenetic tree analysis of YABBY gene family members in sunflower ★,●,▲ and ■indicate Arabidopsis, sunflower, Chrysanthemum morifolium and Lactuca sativa respectively

图3 YABBY家族成员的基因结构和保守基序 A:YABBY家族成员的保守基序图;B:YABBY家族成员的基因结构图;C:YABBY家族成员核心保守结构域氨基酸列比图
Fig. 3 Gene structure and conserved motifs for YABBY family members A: Mapping of conserved motifs for YABBY family members. B: Mapping of gene structure for YABBY family members. C: Amino acid sequence comparison of core conserved domain of YABBY family members
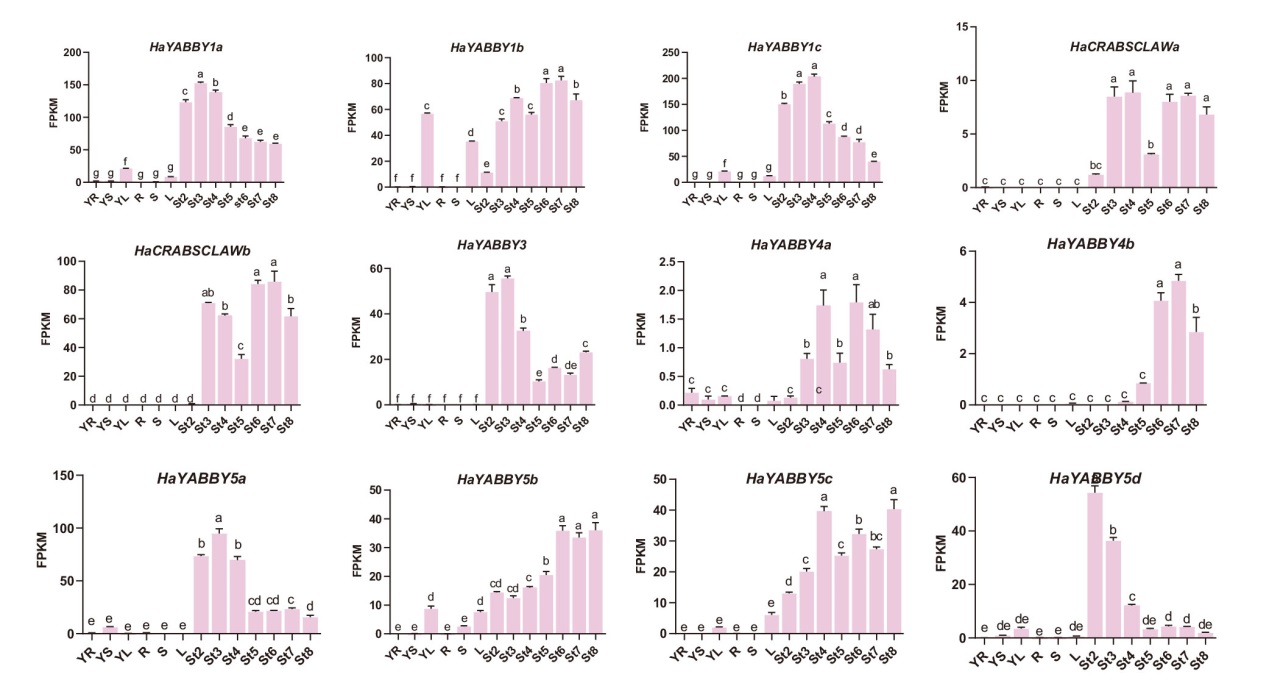
图4 YABBY基因在野生型(WT)向日葵中的组织表达模式分析 FPKM为YABBY基因家族转录丰度;表达组织:YR:幼根;YS:幼茎;YL:幼叶;R:根;S:茎;L:叶;表达时期:St2:花原基起始期;St3:花器官开始形成期;St4:减数分裂期;St5:花粉成熟期;St6:小花开放期;St7:果实灌浆期;St8:种子饱满期;小写字母表示差异显著(P<0.05);下同
Fig. 4 Analysis of tissue expression pattern of YABBY gene in WT sunflower FPKM refers to transcriptional abundance of YABBY gene family. Tissues: YR: young root; YS: young stem; YL: young leaf; R: root; S: stem; L: leaf. Expression period: St2: flower primordium initiation stage; St3: flower organ formation stage; St4: meiosis stage; St5: pollen maturation stage; St6: florets opening stage; St7: fruit filling stage and St8 seed full stage. The lowercase letter indicates significant difference at P<0.05 level. The same below

图5 YABBY基因在野生型(WT)向日葵和cb1突变体中的差异表达分析 A:野生型和cb1突变体花序的表型特征;B:野生型和cb1突变体花序st2-st8表型特征;C:YABBY基因在WT和cb1花序St2-St8的相对表达量分析。所用数据为平均值±SD(n=3);*:显著差异(P<0.05);**:极显著差异(P<0.01)
Fig. 5 Differential expression analysis of YABBY gene in WT sunflower and cb1 mutant A: Phenotypic characteristics of WT sunflower and cb1 mutant inflorescence. B: Phenotypic characteristics of inflorescences in WT and cb1 mutant sunflower from St2-St8. C: Relative expression of YABBY gene in WT and cb1 during inflorescence development(st2-st8). All data are mean ±SD(n=3). *: Significant difference(P<0.05); **: extremely significant difference(P<0.01)
| [1] |
Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development[J]. Trends Genet, 2010, 26(12): 519-527.
doi: 10.1016/j.tig.2010.09.001 pmid: 20947199 |
| [2] |
Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, et al. DNA-binding specificities of plant transcription factors and their potential to define target genes[J]. Proc Natl Acad Sci USA, 2014, 111(6): 2367-2372.
doi: 10.1073/pnas.1316278111 pmid: 24477691 |
| [3] |
Porto MS, Pinheiro MPN, Batista VGL, et al. Plant promoters: an approach of structure and function[J]. Mol Biotechnol, 2014, 56(1): 38-49.
pmid: 24122284 |
| [4] |
Udvardi MK, Kakar K, Wandrey M, et al. Legume transcription factors: global regulators of plant development and response to the environment[J]. Plant Physiol, 2007, 144(2): 538-549.
doi: 10.1104/pp.107.098061 pmid: 17556517 |
| [5] | 刁志娟. 水稻三个花器官发育关键基因和一个SUPERMAN-like基因的功能和表达分析[D]. 福州: 福建农林大学, 2011. |
| Diao ZJ. Functional and expressional analysis of three pivotal genes for floral organ development and a SUPERMAN-like gene in rice(Oryza sativa L.)[D]. Fuzhou: Fujian Agriculture and Forestry University, 2011. | |
| [6] | Nam J, DePamphilis CW, Ma H, et al. Antiquity and evolution of the MADS-box gene family controlling flower development in plants[J]. Mol Biol Evol, 2003, 20(9): 1435-1447. |
| [7] | Mantegazza O, Gregis V, Mendes MA, et al. Analysis of the Arabidopsis REM gene family predicts functions during flower development[J]. Ann Bot, 2014, 114(7): 1507-1515. |
| [8] | Costanzo E, Trehin C, Vandenbussche M. The role of WOX genes in flower development[J]. Ann Bot, 2014, 114(7): 1545-1553. |
| [9] | Viola IL, Gonzalez DH. TCP transcription factors in plant reproductive development: juggling multiple roles[J]. Biomolecules, 2023, 13(5): 750. |
| [10] | Yamada T, Yokota S, Hirayama Y, et al. Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms[J]. Plant J, 2011, 67(1): 26-36. |
| [11] | Bartholmes C, Hidalgo O, Gleissberg S. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica(Papaveraceae)[J]. Plant Biol, 2012, 14(1): 11-23. |
| [12] | Finet C, Floyd SK, Conway SJ, et al. Evolution of the YABBY gene family in seed plants[J]. Evol Dev, 2016, 18(2): 116-126. |
| [13] |
Zhang TP, Li CY, Li DX, et al. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants[J]. J Plant Res, 2020, 133(6): 751-763.
doi: 10.1007/s10265-020-01227-7 pmid: 33033876 |
| [14] | Romanova MA, Maksimova AI, Pawlowski K, et al. YABBY genes in the development and evolution of land plants[J]. Int J Mol Sci, 2021, 22(8): 4139. |
| [15] |
Siegfried KR, Eshed Y, Baum SF, et al. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis[J]. Development, 1999, 126(18): 4117-4128.
doi: 10.1242/dev.126.18.4117 pmid: 10457020 |
| [16] | Sawa S, Watanabe K, Goto K, et al. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains[J]. Genes Dev, 1999, 13(9): 1079-1088. |
| [17] |
Bowman JL. The YABBY gene family and abaxial cell fate[J]. Curr Opin Plant Biol, 2000, 3(1): 17-22.
doi: 10.1016/s1369-5266(99)00035-7 pmid: 10679447 |
| [18] | Lee JY, Baum SF, Oh SH, et al. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade[J]. Development, 2005, 132(22): 5021-5032. |
| [19] | Yamada T, Ito M, Kato M. YABBY2-homologue expression in lateral organs of Amborella trichopoda(Amborellaceae)[J]. Int J Plant Sci, 2004, 165(6): 917-924. |
| [20] |
Sawa S, Ito T, Shimura Y, et al. FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems[J]. Plant Cell, 1999, 11(1): 69-86.
pmid: 9878633 |
| [21] |
Bowman JL, Smyth DR. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains[J]. Development, 1999, 126(11): 2387-2396.
doi: 10.1242/dev.126.11.2387 pmid: 10225998 |
| [22] | Villanueva JM, Broadhvest J, Hauser BA, et al. INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules[J]. Genes Dev, 1999, 13(23): 3160-3169. |
| [23] | Eshed Y, Izhaki A, Baum SF, et al. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities[J]. Development, 2004, 131(12): 2997-3006. |
| [24] | Sarojam R, Sappl PG, Goldshmidt A, et al. Differentiating Arabidopsis shoots from leaves by combined YABBY activities[J]. Plant Cell, 2010, 22(7): 2113-2130. |
| [25] |
Chen Q, Atkinson A, Otsuga D, et al. The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation[J]. Development, 1999, 126(12): 2715-2726.
doi: 10.1242/dev.126.12.2715 pmid: 10331982 |
| [26] |
Lugassi N, Nakayama N, Bochnik R, et al. A novel allele of FILAMENTOUS FLOWER reveals new insights on the link between inflorescence and floral meristem organization and flower morphogenesis[J]. BMC Plant Biol, 2010, 10: 131.
doi: 10.1186/1471-2229-10-131 pmid: 20584289 |
| [27] | Fourquin C, Vinauger-Douard M, Fogliani B, et al. Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms[J]. Proc Natl Acad Sci U S A, 2005, 102(12): 4649-4654. |
| [28] |
Yamaguchi T, Nagasawa N, Kawasaki S, et al. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa[J]. Plant Cell, 2004, 16(2): 500-509.
doi: 10.1105/tpc.018044 pmid: 14729915 |
| [29] |
Ishikawa M, Ohmori Y, Tanaka W, et al. The spatial expression patterns of DROOPING LEAF orthologs suggest a conserved function in grasses[J]. Genes Genet Syst, 2009, 84(2): 137-146.
pmid: 19556707 |
| [30] | Orashakova S, Lange M, Lange S, et al. The CRABS CLAW ortholog from California poppy(Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation[J]. Plant J, 2009, 58(4): 682-693. |
| [31] |
Balasubramanian S, Schneitz K. NOZZLE links proximal-distal and adaxial-abaxial pattern formation during ovule development in Arabidopsis thaliana[J]. Development, 2002, 129(18): 4291-4300.
doi: 10.1242/dev.129.18.4291 pmid: 12183381 |
| [32] |
Meister RJ, Kotow LM, Gasser CS. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments[J]. Development, 2002, 129(18): 4281-4289.
doi: 10.1242/dev.129.18.4281 pmid: 12183380 |
| [33] | di Rienzo V, Imanifard Z, Mascio I, et al. Functional conservation of the grapevine candidate gene INNER NO OUTER for ovule development and seed formation[J]. Hortic Res, 2021, 8(1): 29. |
| [34] | Tanaka W, Toriba T, Ohmori Y, et al. The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet[J]. Plant Cell, 2012, 24(1): 80-95. |
| [35] | Ali Buttar Z, Yang Y, Sharif R, et al. Genome wide identification, characterization, and expression analysis of YABBY-gene family in wheat (Triticum aestivum L.)[J]. Agronomy, 2020, 10(8): 1189. |
| [36] | Chen YY, Hsiao YY, Chang SB, et al. Genome-wide identification of YABBY genes in Orchidaceae and their expression patterns in Phalaenopsis orchid[J]. Genes, 2020, 11(9): 955. |
| [37] |
Cong B, Barrero LS, Tanksley SD. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication[J]. Nat Genet, 2008, 40(6): 800-804.
doi: 10.1038/ng.144 pmid: 18469814 |
| [38] |
Li Z, Zhang ZH, Yan PC, et al. RNA-Seq improves annotation of protein-coding genes in the cucumber genome[J]. BMC Genomics, 2011, 12: 540.
doi: 10.1186/1471-2164-12-540 pmid: 22047402 |
| [39] |
Zhang X, Ding L, Song AP, et al. DWARF AND ROBUST PLANT regulates plant height via modulating gibberellin biosynthesis in chrysanthemum[J]. Plant Physiol, 2022, 190(4): 2484-2500.
doi: 10.1093/plphys/kiac437 pmid: 36214637 |
| [40] |
Gross T, Broholm S, Becker A. CRABS CLAW acts as a bifunctional transcription factor in flower development[J]. Front Plant Sci, 2018, 9: 835.
doi: 10.3389/fpls.2018.00835 pmid: 29973943 |
| [41] | Yang ZE, Gong Q, Wang LL, et al. Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses[J]. Front Genet, 2018, 9: 33. |
| [42] | Wils CR, Kaufmann K. Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana[J]. Biochim Biophys Acta Gene Regul Mech, 2017, 1860(1): 95-105. |
| [43] | Lee DY, An G. Two AP2 family genes, SUPERNUMERARY BRACT(SNB)and OsINDETERMINATE SPIKELET 1(OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice[J]. Plant J, 2012, 69(3): 445-461. |
| [44] | Melzer S, Lens F, Gennen J, et al. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana[J]. Nat Genet, 2008, 40(12): 1489-1492. |
| [45] | Jiu ST, Zhang YP, Han P, et al. Genome-wide identification and expression analysis of VviYABs family reveal its potential functions in the developmental switch and stresses response during grapevine development[J]. Front Genet, 2022, 12: 762221. |
| [46] | Mäkilä R, Wybouw B, Smetana O, et al. Gibberellins promote polar auxin transport to regulate stem cell fate decisions in cambium[J]. Nat Plants, 2023, 9(4): 631-644. |
| [47] | Freire-Rios A, Tanaka K, Crespo I, et al. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis[J]. Proc Natl Acad Sci U S A, 2020, 117(39): 24557-24566. |
| [48] |
Vaddepalli P, Scholz S, Schneitz K. Pattern formation during early floral development[J]. Curr Opin Genet Dev, 2015, 32: 16-23.
doi: 10.1016/j.gde.2015.01.001 pmid: 25687790 |
| [49] | Dai MQ, Hu YF, Zhao Y, et al. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development[J]. Plant Physiol, 2007, 144(1): 380-390. |
| [50] | Siefers N, Dang KK, Kumimoto RW, et al. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity[J]. Plant Physiol, 2009, 149(2): 625-641. |
| [1] | 杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141. |
| [2] | 胡雅丹, 伍国强, 刘晨, 魏明. MYB转录因子在调控植物响应逆境胁迫中的作用[J]. 生物技术通报, 2024, 40(6): 5-22. |
| [3] | 秦健, 李振月, 何浪, 李俊玲, 张昊, 杜荣. 肌源性细胞分化的单细胞转录谱变化及细胞间通讯分析[J]. 生物技术通报, 2024, 40(6): 330-342. |
| [4] | 陈春林, 李白雪, 李金玲, 杜清洁, 李猛, 肖怀娟. 甜瓜CmEPF基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(4): 130-138. |
| [5] | 陈强, 黄馨慧, 张峥, 张冲, 柳叶飞. 褪黑素对薄皮甜瓜采后软化和乙烯合成的影响[J]. 生物技术通报, 2024, 40(4): 139-147. |
| [6] | 张玉, 石磊, 巩檑, 聂峰杰, 杨江伟, 刘璇, 杨文静, 张国辉, 颉瑞霞, 张丽. 马铃薯WOX基因家族的鉴定及在离体再生和非生物胁迫中的表达分析[J]. 生物技术通报, 2024, 40(3): 170-180. |
| [7] | 吴星星, 洪海波, 甘志承, 李瑞宁, 黄先忠. 辣椒CaPI的克隆与功能分析[J]. 生物技术通报, 2024, 40(3): 193-201. |
| [8] | 江林琪, 赵佳莹, 郑飞雄, 姚馨怡, 李效贤, 俞振明. 铁皮石斛14-3-3基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(3): 229-241. |
| [9] | 吴圳, 张明英, 闫锋, 李依民, 高静, 颜永刚, 张岗. 掌叶大黄(Rheum palmatum L.)WRKY基因家族鉴定与分析[J]. 生物技术通报, 2024, 40(1): 250-261. |
| [10] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [11] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [12] | 刘奎, 李兴芬, 杨沛欣, 仲昭晨, 曹一博, 张凌云. 青杄转录共激活因子PwMBF1c的功能研究与验证[J]. 生物技术通报, 2023, 39(5): 205-216. |
| [13] | 赖瑞联, 冯新, 高敏霞, 路喻丹, 刘晓驰, 吴如健, 陈义挺. 猕猴桃过氧化氢酶基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2023, 39(4): 136-147. |
| [14] | 郭三保, 宋美玲, 李灵心, 尧子钊, 桂明明, 黄胜和. 斑地锦查尔酮合酶基因及启动子的克隆与分析[J]. 生物技术通报, 2023, 39(4): 148-156. |
| [15] | 陈强, 邹明康, 宋家敏, 张冲, 吴隆坤. 甜瓜LBD基因家族的鉴定和果实发育进程中的表达分析[J]. 生物技术通报, 2023, 39(3): 176-183. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||