








生物技术通报 ›› 2024, Vol. 40 ›› Issue (8): 74-82.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0201
崔原瑗( ), 王昭懿, 白双宇, 任毓昭, 豆飞飞, 刘彩霞, 刘凤楼, 王掌军, 李清峰(
), 王昭懿, 白双宇, 任毓昭, 豆飞飞, 刘彩霞, 刘凤楼, 王掌军, 李清峰( )
)
收稿日期:2024-03-14
出版日期:2024-08-26
发布日期:2024-07-30
通讯作者:
李清峰,男,博士,副教授,研究方向:麦类作物遗传育种;E-mail: liqingfeng2017@nxu.edu.cn作者简介:崔原瑗,女,硕士研究生,研究方向:麦类作物遗传育种;E-mail: 18695124679@163.com
基金资助:
CUI Yuan-yuan( ), WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng(
), WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng( )
)
Received:2024-03-14
Published:2024-08-26
Online:2024-07-30
摘要:
【目的】非特异性磷脂酶C(non-specific phospholipase C, NPC)是磷脂酶C的一种,在植物生长发育和响应非生物胁迫等方面发挥重要作用。在大麦全基因组范围内鉴定NPC基因家族成员,分析相关基因表达特点,为大麦NPC基因功能研究及遗传改良提供理论依据。【方法】基于EnsemblPlants数据库收录的大麦全基因组数据,利用生物信息学的方法对大麦NPC基因家族进行鉴定,并对大麦NPC基因家族的理化特性、亚细胞定位、系统进化关系、基因结构、蛋白结构域和启动子顺势元件等进行分析。利用转录组数据和实时荧光定量PCR分析HvNPC基因在不同组织以及苗期在低温、干旱、盐胁迫下的表达特征。【结果】在大麦基因组中,共鉴定了5个非特异性磷脂酶C基因家族成员,其编码蛋白与其他植物的NPC基因结构相似,均具有Phosphoesterase结构域;HvNPC蛋白序列长度为477-553 aa,等电点为5.22-8.83,分子量为53.12-61.33 kD;亚细胞定位预测HvNPC基因定位于叶绿体、液泡膜、细胞质中;HvNPC启动子区含有大量与非生物胁迫有关的顺势作用元件。实时荧光定量PCR分析证实HvNPC基因受干旱、高盐及低温胁迫诱导表达,其中HvNPC2、HvNPC3和HvNPC5受低温和干旱胁迫诱导表达;HvNPC3和HvNPC5受盐胁迫诱导表达。【结论】在大麦基因组中共鉴定5个HvNPC基因,HvNPC基因成员具有器官表达特异性,并且不同基因对非生物胁迫响应不同。
崔原瑗, 王昭懿, 白双宇, 任毓昭, 豆飞飞, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 大麦非特异性磷脂酶C基因家族全基因组鉴定及苗期胁迫表达分析[J]. 生物技术通报, 2024, 40(8): 74-82.
CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage[J]. Biotechnology Bulletin, 2024, 40(8): 74-82.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 退火温度Annealing temperature/℃ |
|---|---|---|
| HvNPC1-F | AACAAGCCCCCATCACCG | 58.9 |
| HvNPC1-R | CCACCTTGAAGGCGTCGT | 58.6 |
| HvNPC2-F | ACTACCAGTACCCGCCGT | 58.9 |
| HvNPC2-R | ACCACGTAGTTGGGCAGC | 58.6 |
| HvNPC3-F | GACGTGCTGGACCAGGTC | 59.4 |
| HvNPC3-R | CGGAGGAGAGCAGGGAGT | 59.8 |
| HvNPC4-F | GATGCCGACGAGGATCCG | 59.2 |
| HvNPC4-R | ATCCCCGTTCAGCACTGC | 58.8 |
| HvNPC5-F | CGTCTTCGACCGCTGGTT | 58.4 |
| HvNPC5-R | CGTTGAAGGTGAGCCCGT | 58.6 |
| HvActin-F | CAGGTATCGCTGACCGTATGAG | 57.5 |
| HvActin-R | TGGAAAGTGCTGAGTGAGGCT | 58.9 |
表1 RT-qPCR 引物
Table 1 Primers used for RT-qPCR
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 退火温度Annealing temperature/℃ |
|---|---|---|
| HvNPC1-F | AACAAGCCCCCATCACCG | 58.9 |
| HvNPC1-R | CCACCTTGAAGGCGTCGT | 58.6 |
| HvNPC2-F | ACTACCAGTACCCGCCGT | 58.9 |
| HvNPC2-R | ACCACGTAGTTGGGCAGC | 58.6 |
| HvNPC3-F | GACGTGCTGGACCAGGTC | 59.4 |
| HvNPC3-R | CGGAGGAGAGCAGGGAGT | 59.8 |
| HvNPC4-F | GATGCCGACGAGGATCCG | 59.2 |
| HvNPC4-R | ATCCCCGTTCAGCACTGC | 58.8 |
| HvNPC5-F | CGTCTTCGACCGCTGGTT | 58.4 |
| HvNPC5-R | CGTTGAAGGTGAGCCCGT | 58.6 |
| HvActin-F | CAGGTATCGCTGACCGTATGAG | 57.5 |
| HvActin-R | TGGAAAGTGCTGAGTGAGGCT | 58.9 |
| 基因名称 Gene name | 基因ID Transcript ID | 物理位置 Physical position | 相对分子量 Molecular weight/kD | 等电点 pI | 亲水性总平均值 Grand average of hydrophilicity | 蛋白长度 Protein length/aa | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| HvNPC1 | HORVU.MOREX.r3.7HG0752530.1 | 7H:631001620-631003350 | 53.12 | 5.22 | -0.317 | 477 | 细胞质 |
| HvNPC2 | HORVU.MOREX.r3.1HG0085590.1 | 1H:496122549-496124474 | 61.33 | 6.53 | -0.364 | 553 | 叶绿体 |
| HvNPC3 | HORVU.MOREX.r3.3HG0232030.1 | 3H:28367740-28369602 | 58.77 | 8.83 | -0.204 | 532 | 叶绿体 |
| HvNPC4 | HORVU.MOREX.r3.3HG0322080.1 | 3H:600541735-600546962 | 59.07 | 5.98 | -0.424 | 533 | 液泡膜 |
| HvNPC5 | HORVU.MOREX.r3.5HG0527960.1 | 5H:566994044-566997667 | 60.76 | 6.80 | -0.367 | 545 | 叶绿体 |
表2 大麦NPC基因家族成员基本理化性质
Table 2 Basic physico-chemical properties of NPC family in barley(Hordeum vulgare L)
| 基因名称 Gene name | 基因ID Transcript ID | 物理位置 Physical position | 相对分子量 Molecular weight/kD | 等电点 pI | 亲水性总平均值 Grand average of hydrophilicity | 蛋白长度 Protein length/aa | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| HvNPC1 | HORVU.MOREX.r3.7HG0752530.1 | 7H:631001620-631003350 | 53.12 | 5.22 | -0.317 | 477 | 细胞质 |
| HvNPC2 | HORVU.MOREX.r3.1HG0085590.1 | 1H:496122549-496124474 | 61.33 | 6.53 | -0.364 | 553 | 叶绿体 |
| HvNPC3 | HORVU.MOREX.r3.3HG0232030.1 | 3H:28367740-28369602 | 58.77 | 8.83 | -0.204 | 532 | 叶绿体 |
| HvNPC4 | HORVU.MOREX.r3.3HG0322080.1 | 3H:600541735-600546962 | 59.07 | 5.98 | -0.424 | 533 | 液泡膜 |
| HvNPC5 | HORVU.MOREX.r3.5HG0527960.1 | 5H:566994044-566997667 | 60.76 | 6.80 | -0.367 | 545 | 叶绿体 |

图2 大麦NPC基因家族的结构特征 A:紫色条表示外显子,黑线表示内含子,灰色条表示第灰色条表示非翻译区(UTR);B:黄色条表示特征域phosphoesterase(PF04185)
Fig. 2 Structural features of NPC gene family in barely A: Purple bars indicate the exon, lines indicate the intron while grey color bars indicate the un-translated region(UTR). B: The yellow bar indicates the signature domain phosphoesterase(PF04185)
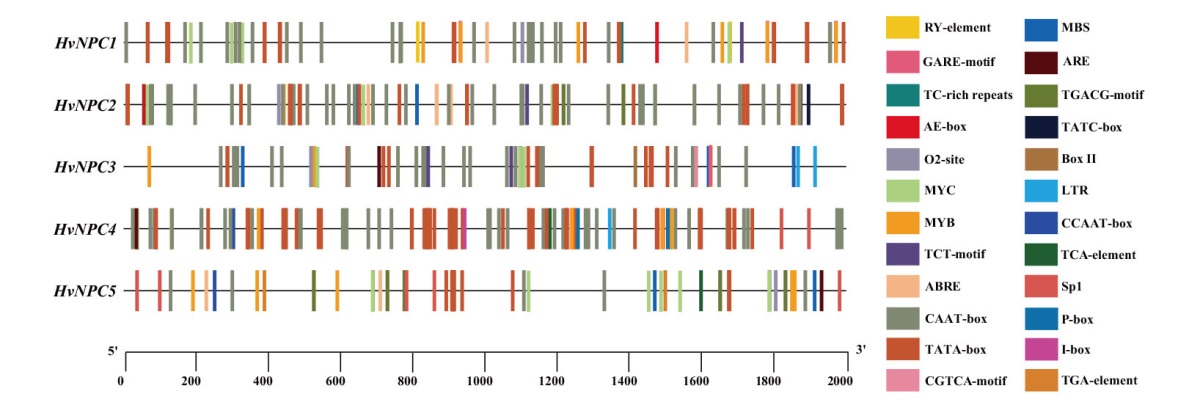
图3 HvNPC基因家族各成员启动子区顺式作用元件 ABRE:脱落酸响应元件;CGTCA-motif、TGACG-motif:茉莉酸甲酯响应元件;GARE-motif、P-box、TATC-box:赤霉素响应元件;TGA-element:生长素响应元件;TCA-element:水杨酸响应元件;MBS、MYC、MYB:干旱胁迫响应元件;TC-rich:防御和胁迫响应元件;LTR:低温胁迫响应元件;ARE:厌氧诱导响应元件;O2-site:玉米醇溶蛋白代谢调控元件;RY-element:种子特异性调控元件;Sp1、TCT-motif、AE-box、BoxII、I-box:光响应元件
Fig. 3 Cis-acting elements in the promoters of HvNPC gene family members ABRE: Abscisic acid responsive element. CGTCA-motif, TGACG-motif: MeJA responsive element. GARE-motif, P-box, TATC-box: Gibberellin responsive element. TGA-element: Auxin responsive element. TCA-element: Salicylic acid responsive element. MBS, MYC, MYB: Drought responsive element. TC-rich: Defense and stress responsive element. LTR: Low-temperature responsive element. ARE: Essential for the anaerobic responsive element. O2-site: Zein metabolism regulatory element. RY-element: Seed-specific regulatory element. Sp1, TCT-motif, AE-box, BoxII, I-box: Light responsive element
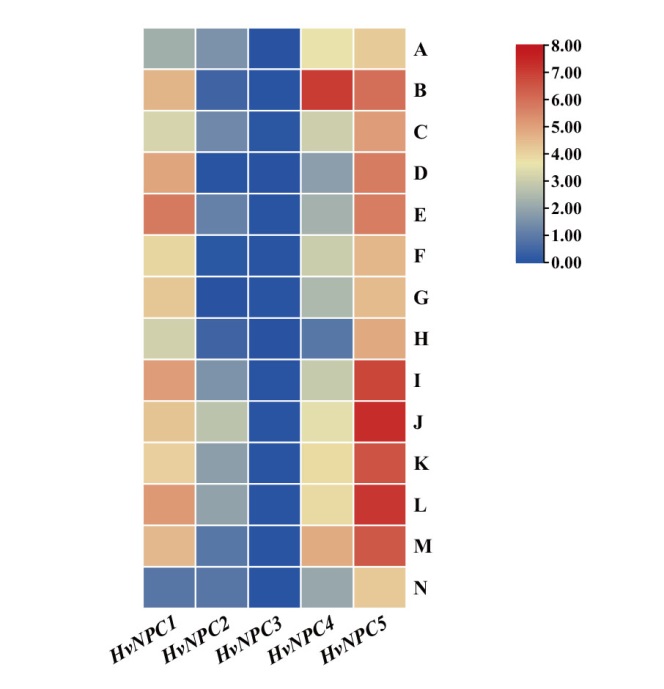
图4 大麦NPC基因在14个组织器官中的表达谱 A:胚(萌发后4 d);B:种子根(茎长10 cm);C:茎(10 cm);D:黄化幼苗(黑暗条件下10 d);E:分蘖(6叶期,第3节);F:根(4周); G:表皮(4周);H:花序(5 mm);I:花序(1-1.5 cm);J:花轴(5周);K:外稃(6周);L:浆片(6周);M:内稃(6周);N:衰老叶片(2周)
Fig. 4 Expression profile of NPC gene in 14 tissues and organs of barley A: Embryo(4 d after germination). B: Roots from the seedlings(shoot length 10 cm). C: Shoots(10 cm). D: Etiolated seedling (10 d old at dark). E: Developing tillers(six leaf stge, 3rd internode). F: Root(4 weeks). G: Epidermis(4 weeks). H: Florescences(5 mm). I: Florescences(1-1.5 cm). J: Rachis(5 weeks). K: Lemma(6 weeks). L: Lodicule(6 weeks). M: Palea(6 weeks). N: Senescing leaf(2 weeks)
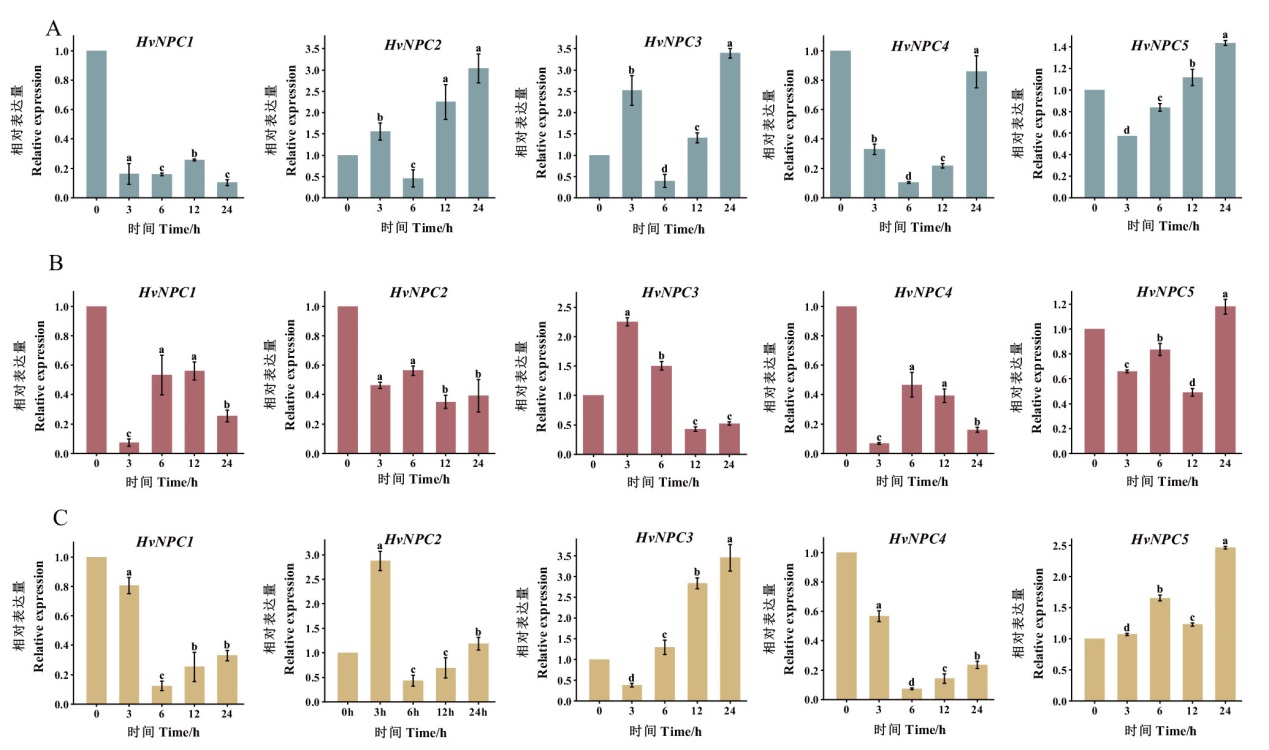
图5 低温、氯化钠、干旱处理下HvNPCs基因的表达模式 A:4℃处理下HvNPCs的基因表达谱;B:NaCl处理下HvNPCs的基因表达谱;C:PEG处理下HvNPCs的基因表达谱;不同小写字母代表显著差异(P<0.05)
Fig. 5 Relative expression pattern of HvNPCs genes under low temperature, NaCl, and drought treatments A: Gene expression profiles of HvNPCs under 4℃ treatment. B: Gene expression profiles of HvNPCs under NaCl treatment. C: Gene expression profiles of HvNPCs under PEG treatment. Different lower letters indicate significant differences(P<0.05)
| [1] | Zhou J, Wang JJ, Bi YF, et al. Overexpression of PtSOS2 enhances salt tolerance in transgenic poplars[J]. Plant Mol Biol Report, 2014, 32(1): 185-197. |
| [2] |
Wang WX, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance[J]. Planta, 2003, 218(1): 1-14.
doi: 10.1007/s00425-003-1105-5 pmid: 14513379 |
| [3] |
Testerink C, Munnik T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants[J]. J Exp Bot, 2011, 62(7): 2349-2361.
doi: 10.1093/jxb/err079 pmid: 21430291 |
| [4] | Ali U, Lu SP, Fadlalla T, et al. The functions of phospholipases and their hydrolysis products in plant growth, development and stress responses[J]. Prog Lipid Res, 2022, 86: 101158. |
| [5] |
Nakamura Y, Awai K, Masuda T, et al. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis[J]. J Biol Chem, 2005, 280(9): 7469-7476.
doi: 10.1074/jbc.M408799200 pmid: 15618226 |
| [6] | Chen WH, Taylor MC, Barrow RA, et al. Loss of phosphoethanolamine N-methyltransferases abolishes phosphatidylcholine synthesis and is lethal[J]. Plant Physiol, 2019, 179(1): 124-142. |
| [7] | Mou ZL, Wang XQ, Fu ZM, et al. Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis[J]. Plant Cell, 2002, 14(9): 2031-2043. |
| [8] |
Pokotylo I, Pejchar P, Potocký M, et al. The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling[J]. Prog Lipid Res, 2013, 52(1): 62-79.
doi: 10.1016/j.plipres.2012.09.001 pmid: 23089468 |
| [9] |
Krčková Z, Brouzdová J, Daněk M, et al. Arabidopsis non-specific phospholipase C1: characterization and its involvement in response to heat stress[J]. Front Plant Sci, 2015, 6: 928.
doi: 10.3389/fpls.2015.00928 pmid: 26581502 |
| [10] | Krcková Z, Kocourková D, Danek M, et al. The Arabidopsis thaliana non-specific phospholipase C2 is involved in the response to Pseudomonas syringae attack[J]. Ann Bot, 2018, 121(2): 297-310. |
| [11] | Peters C, Li MY, Narasimhan R, et al. Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis[J]. Plant Cell, 2010, 22(8): 2642-2659. |
| [12] | Pejchar P, Potocký M, Krčková Z, et al. Non-specific phospholipase C4 mediates response to aluminum toxicity in Arabidopsis thaliana[J]. Front Plant Sci, 2015, 6: 66. |
| [13] | Peters C, Kim SC, Devaiah S, et al. Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis[J]. Plant Cell Environ, 2014, 37(9): 2002-2013. |
| [14] |
Cai GQ, Fan CC, Liu S, et al. Nonspecific phospholipase C6 increases seed oil production in oilseed Brassicaceae plants[J]. New Phytol, 2020, 226(4): 1055-1073.
doi: 10.1111/nph.16473 pmid: 32176333 |
| [15] | She KJ, Pan WQ, Yan Y, et al. Genome-wide identification, evolution and expressional analysis of OSCA gene family in barley(Hordeum vulgare L.)[J]. Int J Mol Sci, 2022, 23(21): 13027. |
| [16] |
Wang G, Ryu S, Wang X. Plant phospholipases: an overview[J]. Methods Mol Biol, 2012, 861: 123-137.
doi: 10.1007/978-1-61779-600-5_8 pmid: 22426716 |
| [17] | Wang XG, Liu Y, Li Z, et al. Genome-wide identification and expression profile analysis of the phospholipase C gene family in wheat(Triticum aestivum L.)[J]. Plants, 2020, 9(7): 885. |
| [18] | Singh A, Kanwar P, Pandey A, et al. Comprehensive genomic analysis and expression profiling of phospholipase C gene family during abiotic stresses and development in rice[J]. PLoS One, 2013, 8(4): e62494. |
| [19] | Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal[J]. Nucleic Acids Res, 2012, 40(Web Server issue): W597-W603. |
| [20] |
Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Mol Biol Evol, 2011, 28(10): 2731-2739.
doi: 10.1093/molbev/msr121 pmid: 21546353 |
| [21] |
Letunic I, Bork P. Interactive Tree Of Life(iTOL)v4: recent updates and new developments[J]. Nucleic Acids Res, 2019, 47(W1): W256-W259.
doi: 10.1093/nar/gkz239 |
| [22] | Chen CJ, Chen H, He YH, et al. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface[J]. bioRxiv, 2018. DOI: 10.1101/289660. |
| [23] |
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327.
doi: 10.1093/nar/30.1.325 pmid: 11752327 |
| [24] | Li TT, Pan WQ, Yuan YY, et al. Identification, characterization, and expression profile analysis of the mTERF gene family and its role in the response to abiotic stress in barley(Hordeum vulgare L.)[J]. Front Plant Sci, 2021, 12: 684619. |
| [25] |
Zhang B, Wang YM, Liu JY. Genome-wide identification and characterization of phospholipase C gene family in cotton(Gossypium spp.)[J]. Sci China Life Sci, 2018, 61(1): 88-99.
doi: 10.1007/s11427-017-9053-y pmid: 28547583 |
| [26] | Tasma IM, Brendel V, Whitham SA, et al. Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana[J]. Plant Physiol Biochem, 2008, 46(7): 627-637. |
| [27] | Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis[J]. PLoS Biol, 2004, 2(9): E258. |
| [28] | Zhu JT, Zhou YY, Li JL, et al. Genome-wide investigation of the phospholipase C gene family in Zea mays[J]. Front Genet, 2021, 11: 611414. |
| [29] | Song JL, Zhou YH, Zhang JR, et al. Structural, expression and evolutionary analysis of the non-specific phospholipase C gene family in Gossypium hirsutum[J]. BMC Genomics, 2017, 18(1): 979. |
| [30] | Yang D, Liu X, Yin XM, et al. Rice non-specific phospholipase C6 is involved in mesocotyl elongation[J]. Plant Cell Physiol, 2021, 62(6): 985-1000. |
| [1] | 李雨晴, 吴楠, 罗建让. 卵叶牡丹花色苷合成相关基因bHLH的克隆与功能分析[J]. 生物技术通报, 2024, 40(8): 174-185. |
| [2] | 李亦君, 杨小贝, 夏琳, 罗朝鹏, 徐馨, 杨军, 宁黔冀, 武明珠. 烟草NtPRR37基因克隆及功能分析[J]. 生物技术通报, 2024, 40(8): 221-231. |
| [3] | 李勇慧, 鲍星星, 段一珂, 赵运霞, 于相丽, 陈尧, 张延召. 灵宝杜鹃bZIP家族全基因组鉴定及表达特征分析[J]. 生物技术通报, 2024, 40(8): 186-198. |
| [4] | 刘丹丹, 王雷刚, 孙明慧, 焦小雨, 吴琼, 王文杰. 茶树海藻糖-6-磷酸合成酶(TPS)基因家族鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 152-163. |
| [5] | 杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141. |
| [6] | 余纽, 柳帆, 杨锦昌. 油楠SgTPS7的克隆及其在萜类生物合成和非生物胁迫中的功能[J]. 生物技术通报, 2024, 40(8): 164-173. |
| [7] | 周冉, 王兴平, 李彦霞, 罗仍卓么. 金黄色葡萄球菌型乳房炎奶牛乳腺组织的lncRNA差异表达分析[J]. 生物技术通报, 2024, 40(8): 320-328. |
| [8] | 张明亚, 庞胜群, 刘玉东, 苏永峰, 牛博文, 韩琼琼. 番茄FAD基因家族的鉴定与表达分析[J]. 生物技术通报, 2024, 40(7): 150-162. |
| [9] | 臧文蕊, 马明, 砗根, 哈斯阿古拉. 甜瓜BZR转录因子家族基因的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(7): 163-171. |
| [10] | 吴丁洁, 陈盈盈, 徐静, 刘源, 张航, 李瑞丽. 植物赤霉素氧化酶及其功能研究进展[J]. 生物技术通报, 2024, 40(7): 43-54. |
| [11] | 李博静, 郑腊梅, 吴乌云, 高飞, 周宜君. 西蒙得木HSP20基因家族的进化、表达和功能分析[J]. 生物技术通报, 2024, 40(6): 190-202. |
| [12] | 胡雅丹, 伍国强, 刘晨, 魏明. MYB转录因子在调控植物响应逆境胁迫中的作用[J]. 生物技术通报, 2024, 40(6): 5-22. |
| [13] | 阿丽亚·外力, 陈永坤, 克拉热木·克里木江, 王宝庆, 陈凌娜. 核桃SPL基因家族的系统进化和表达分析[J]. 生物技术通报, 2024, 40(6): 180-189. |
| [14] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [15] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||