








生物技术通报 ›› 2025, Vol. 41 ›› Issue (3): 104-111.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0839
• 研究报告 • 上一篇
收稿日期:2024-08-29
出版日期:2025-03-26
发布日期:2025-03-20
通讯作者:
朱新霞,女,博士,研究方向 :植物生物技术;E-mail: 302641316@qq.com作者简介:马利花,女,硕士研究生,研究方向 :生物化学和分子生物学;E-mail: 2271579422@qq.com
基金资助:
MA Li-hua( ), HOU Meng-juan, ZHU Xin-xia(
), HOU Meng-juan, ZHU Xin-xia( )
)
Received:2024-08-29
Published:2025-03-26
Online:2025-03-20
摘要:
目的 分析GhNFD4在陆地棉响应干旱胁迫中的生物学功能,为陆地棉抗逆分子育种提供基因资源和理论依据。 方法 使用实时荧光定量PCR(RT-qPCR)技术检测棉花幼苗在PEG模拟的自然干旱胁迫下的GhNFD4表达模式;利用烟草脆裂病毒(TRV)诱导的基因沉默技术(VIGS)探究GhNFD4在干旱胁迫中的调控作用,克隆GhNFD4干扰序列和构建基因沉默载体,并通过瞬时转染技术侵染棉花幼苗进一步获取GhNFD4沉默植株,观察沉默植株与对照植株在自然失水15 d的表型差异,采用NBT和DAB染色法检测ROS产物的积累,并检测离体叶片的失水率和抗旱相关生理指标等。 结果 GhNFD4的表达受PEG模拟的自然干旱胁迫诱导,并在胁迫12 h表达量达到了顶峰。自然干旱15 d后,与阴性对照植株相比,GhNFD4沉默植株叶片萎蔫较为严重,且叶片在NBT和DAB染色后着色更深,其离体叶片失水率及干旱胁迫后叶片的相对电导率(REL)、丙二醛(MDA)和过氧化氢(H2O2)含量与对照组相比均显著升高,总抗氧化物酶(T-AOC)活性与对照组相比显著下降。 结论 GhNFD4受干旱胁迫诱导,在干旱条件下通过调节抗氧化酶活性参与棉花对干旱胁迫的正向调控,改善棉花对干旱胁迫的抵抗能力。
马利花, 侯梦娟, 朱新霞. 陆地棉GhNFD4在棉花干旱响应中的功能[J]. 生物技术通报, 2025, 41(3): 104-111.
MA Li-hua, HOU Meng-juan, ZHU Xin-xia. Functional Studies of the Gossypium hirsutumGhNFD4 Gene in Response to Drought in Cotton[J]. Biotechnology Bulletin, 2025, 41(3): 104-111.
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途Purpose |
|---|---|---|
| GhUBQ7-F | AGAGGTCGAGTCTTGGGACA | 内参基因Reference gene |
| GhUBQ7-R | GCTTGATCTTCTTGGGCTTG | |
| GhNFD4-F | AAACCTTGTCACTTGGGCGA | 实时荧光定量PCR RT-qPCR |
| GhNFD4-R | CTTTCGGGTACTTCCCTCCG | |
| GhNFD4-EcoRIF | CGGAATTCGACAAAGAGGGCCAAAGATGC | 基因克隆 Gene cloning |
| GhNFD4-BamHIR | CGGGATCCTCACCCCACTCTCCATTTTGT |
表1 本试验所用引物
Table 1 Primers used in this study
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途Purpose |
|---|---|---|
| GhUBQ7-F | AGAGGTCGAGTCTTGGGACA | 内参基因Reference gene |
| GhUBQ7-R | GCTTGATCTTCTTGGGCTTG | |
| GhNFD4-F | AAACCTTGTCACTTGGGCGA | 实时荧光定量PCR RT-qPCR |
| GhNFD4-R | CTTTCGGGTACTTCCCTCCG | |
| GhNFD4-EcoRIF | CGGAATTCGACAAAGAGGGCCAAAGATGC | 基因克隆 Gene cloning |
| GhNFD4-BamHIR | CGGGATCCTCACCCCACTCTCCATTTTGT |
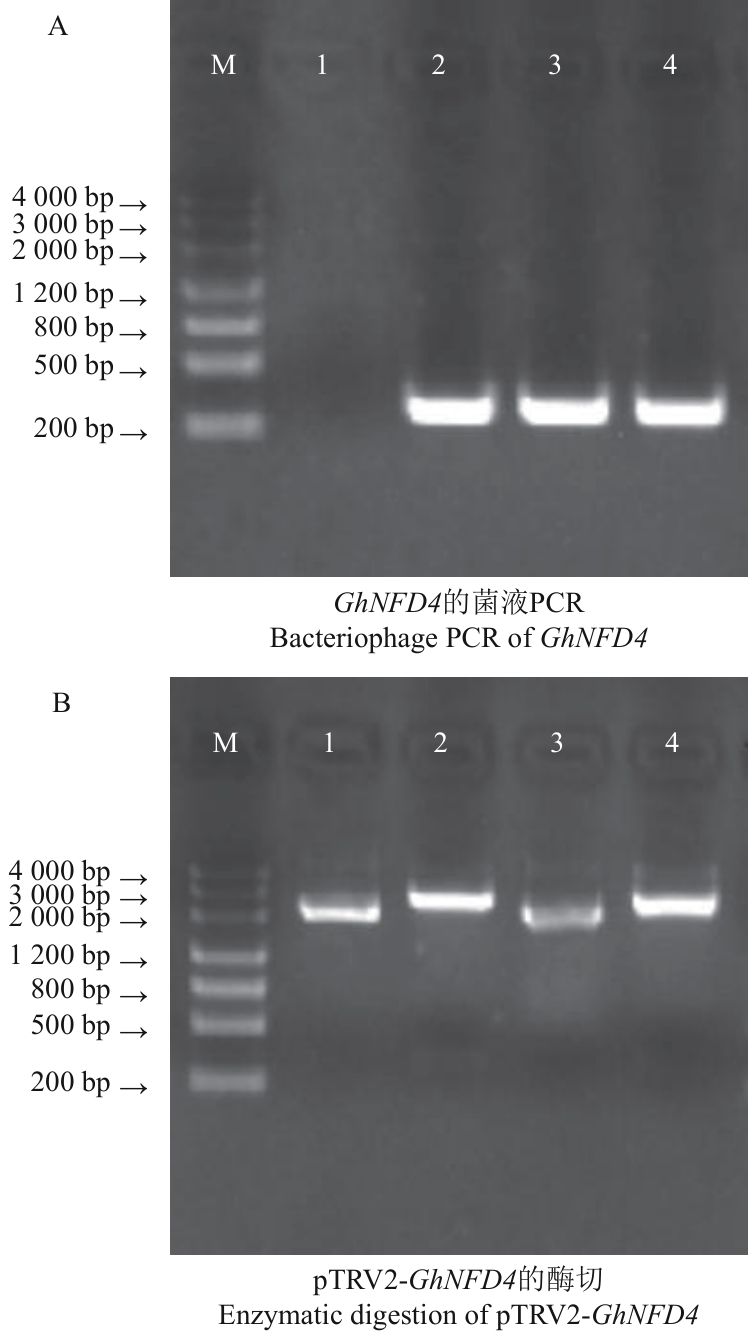
图2 GhNFD4沉默载体的构建A:1:阴性对照;2-4:GhNFD4沉默片段的克隆;B:1-2:pTRV2-GhNFD4(TRV2-NFD4-1)对照及酶切;3-4:pTRV2-GhNFD4(TRV2-NFD4-3)对照及酶切;M:Marker 3000
Fig. 2 Construction of GhNFD4 silencing vectorA: 1: Negative control; 2-4: cloning of GhNFD4 silencing fragment. B: 1-2: pTRV2-GhNFD4 (TRV2-NFD4-1) control and digest; 3-4: pTRV2-GhNFD4 (TRV2-NFD4-3) control and digest. M: Marker 3000
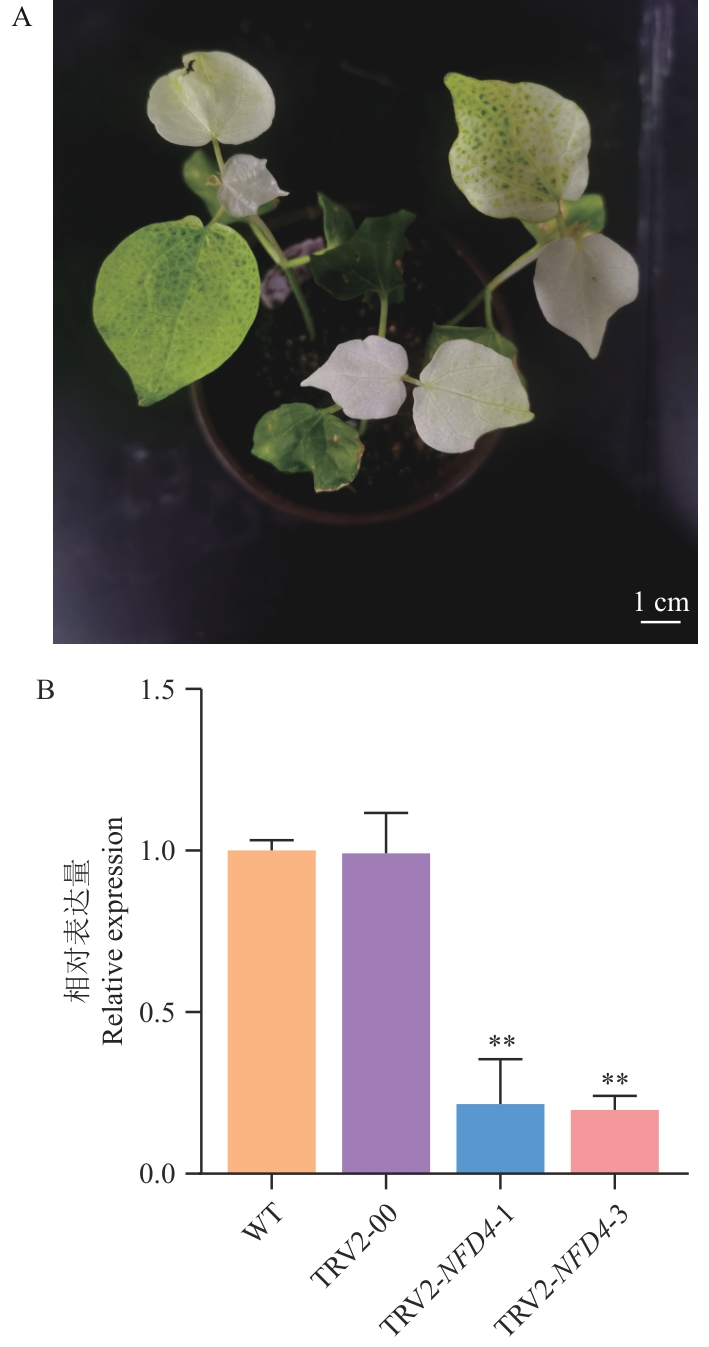
图3 VIGS沉默体系的建立与VIGS沉默效率检测A:注射pTRV1/pTRV2‑GhCLA1的棉花白化表型;B:WT、TRV2-00和VIGS植株(TRV2-GhNFD4-1、TRV2-GhNFD4-3)中GhNFD4的RT-qPCR分析。**P<0.01。下同
Fig. 3 Establishment of VIGS silencing system and detection of VIGS silencing efficiencyA: Cotton albino phenotype injected with pTRV1/pTRV2-GhCLA1. B: RT-qPCR analysis of GhNFD4 in WT, TRV2-00 and VIGS plants (TRV2-GhNFD4-1, TRV2-GhNFD4-3).**P<0.01. The same below
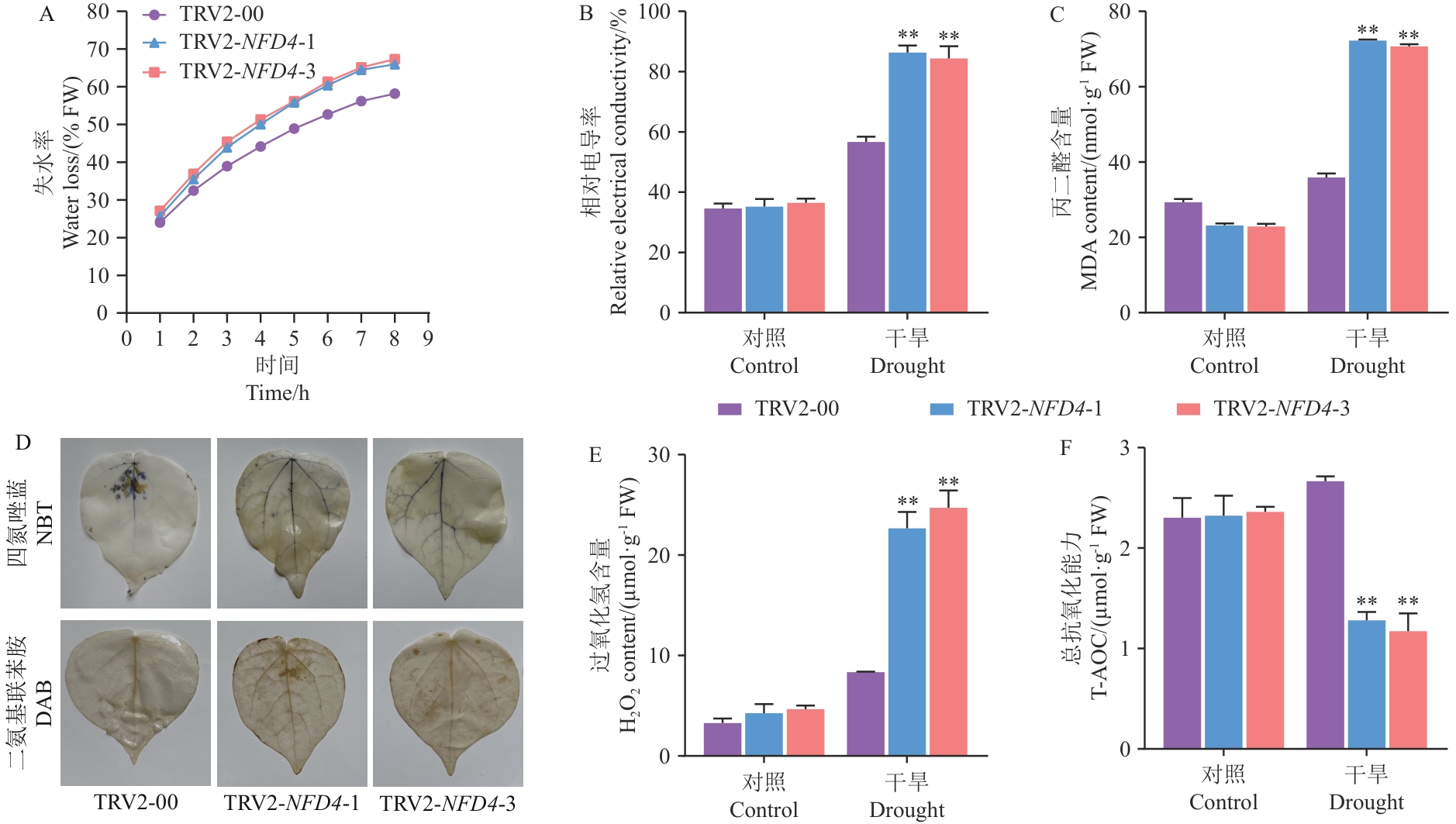
图5 GhNFD4沉默植株在干旱胁迫15 d后的生理指标测定A:离体叶片失水率;B:相对电导率;C:丙二醛含量(MDA);D:干旱胁迫后VIGS和对照植株的DAB和NBT染色;E:过氧化氢含量(H2O2);F:总抗氧化能力(T-AOC)
Fig. 5 Determination for the physiological indices of GhNFD4 gene-silenced plants measured after 15 d of drought stressA: Water loss rate of isolated leaves. B: Relative electrical conductivity. C: Malondialdehyde content (MDA). D: DAB and NBT staining of VIGS and control plants after drought stress. E: Hydrogen peroxide content (H2O2). F: Total antioxidant capacity (T-AOC)
| 1 | Bashir K, Matsui A, Rasheed S, et al. Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress [J]. F1000Res, 2019, 8(F1000 Faculty Rev): 658. |
| 2 | Mahmood U, Li XD, Fan YH, et al. Multi-omics revolution to promote plant breeding efficiency [J]. Front Plant Sci, 2022, 13: 1062952. |
| 3 | Abdelraheem A, Esmaeili N, O’Connell M, et al. Progress and perspective on drought and salt stress tolerance in cotton [J]. Ind Crops Prod, 2019, 130: 118-129. |
| 4 | Ergashovich KA, Azamatovna BZ, Toshtemirovna NU, et al. Ecophysiological effects of water deficiency on cotton varieties [J]. J Crit Rev, 2020, 7(9): 244-246. |
| 5 | Sajid M, Amjid M, Munir H, et al. Comparative analysis of growth and physiological responses of sugarcane elite genotypes to water stress and sandy loam soils [J]. Plants (Basel), 2023, 12(15): 2759. |
| 6 | Fang YJ, Liao KF, Du H, et al. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice [J]. J Exp Bot, 2015, 66(21): 6803-6817. |
| 7 | Deeba F, Pandey AK, Ranjan S, et al. Physiological and proteomic responses of cotton (Gossypium herbaceum L.) to drought stress [J]. Plant Physiol Biochem, 2012, 53: 6-18. |
| 8 | Hussain S, Hussain S, Qadir T, et al. Drought stress in plants: an overview on implications, tolerance mechanisms and agronomic mitigation strategies [J]. Plant Sci Today, 2019, 6(4): 389-402. |
| 9 | Ozturk M, Turkyilmaz Unal B, García-Caparrós P, et al. Osmoregulation and its actions during the drought stress in plants [J]. Physiol Plant, 2021, 172(2): 1321-1335. |
| 10 | Pao SS, Paulsen IT, Jr Saier MH. Major facilitator superfamily [J]. Microbiol Mol Biol Rev, 1998, 62(1): 1-34. |
| 11 | Quistgaard EM, Löw C, Guettou F, et al. Understanding transport by the major facilitator superfamily (MFS): structures pave the way [J]. Nat Rev Mol Cell Biol, 2016, 17(2): 123-132. |
| 12 | Hwang JU, Song WY, Hong D, et al. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle [J]. Mol Plant, 2016, 9(3): 338-355. |
| 13 | Guo RZ, Zhang Q, Qian K, et al. Phosphate-dependent regulation of vacuolar trafficking of OsSPX-MFSs is critical for maintaining intracellular phosphate homeostasis in rice [J]. Mol Plant, 2023, 16(8): 1304-1320. |
| 14 | Julião MHM, Silva SR, Ferro JA, et al. A genomic and transcriptomic overview of mate, abc, and MFS transporters in Citrus sinensis interaction with Xanthomonas citri subsp. citri [J]. Plants (Basel), 2020, 9(6): 794. |
| 15 | Boorer KJ, Loo DD, Wright EM. Steady-state and presteady-state kinetics of the H+/hexose cotransporter (STP1) from Arabidopsis thaliana expressed in Xenopus oocytes [J]. J Biol Chem, 1994, 269(32): 20417-20424. |
| 16 | Cordoba E, Aceves-Zamudio DL, Hernández-Bernal AF, et al. Sugar regulation of sugar transporter protein 1 (stp1) expression in Arabidopsis thaliana [J]. J Exp Bot, 2015, 66(1): 147-159. |
| 17 | Büttner M, Truernit E, Baier K, et al. AtSTP3, a green leaf-specific, low affinity monosaccharide-H+ symporter of Arabidopsis thaliana [J]. Plant Cell Environ, 2000, 23(2): 175-184. |
| 18 | Segonzac C, Boyer JC, Ipotesi E, et al. Nitrate efflux at the root plasma membrane: identification of an Arabidopsis excretion transporter [J]. Plant Cell, 2007, 19(11): 3760-3777. |
| 19 | Liu WW, Sun Q, Wang K, et al. Nitrogen Limitation Adaptation (NLA) is involved in source-to-sink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis [J]. New Phytol, 2017, 214(2): 734-744. |
| 20 | Taochy C, Gaillard I, Ipotesi E, et al. The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress [J]. Plant J, 2015, 83(3): 466-479. |
| 21 | Liu TY, Huang TK, Yang SY, et al. Identification of plant vacuolar transporters mediating phosphate storage [J]. Nat Commun, 2016, 7: 11095. |
| 22 | Lu JY, Wang CL, Wang HY, et al. OsMFS1/OsHOP2 complex participates in rice male and female development [J]. Front Plant Sci, 2020, 11: 518. |
| 23 | Kong H, Hou MJ, Ma B, et al. Calcium-dependent protein kinase GhCDPK4 plays a role in drought and abscisic acid stress responses [J]. Plant Sci, 2023, 332: 111704. |
| 24 | Mandel MA, Feldmann KA, Herrera-Estrella L, et al. CLA1 a novel gene required for chloroplast development, is highly conserved in evolution [J]. Plant J, 1996, 9(5): 649-658. |
| 25 | Wei TL, Wang Y, Xie ZZ, et al. Enhanced ROS scavenging and sugar accumulation contribute to drought tolerance of naturally occurring autotetraploids in Poncirus trifoliata [J]. Plant Biotechnol J, 2019, 17(7): 1394-1407. |
| 26 | Zou LP, Qi DF, Sun JB, et al. Expression of the cassava nitrate transporter NRT2.1 enables Arabidopsis low nitrate tolerance [J]. J Genet, 2019, 98: 74. |
| 27 | Yan HL, Xu WX, Xie JY, et al. Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies [J]. Nat Commun, 2019, 10(1): 2562. |
| 28 | Zhang XM, Feng JJ, Zhao RL, et al. Functional characterization of the GhNRT2.1e gene reveals its significant role in improving nitrogen use efficiency in Gossypium hirsutum [J]. PeerJ, 2023, 11: e15152. |
| 29 | Liu FJ, Cai S, Dai LJ, et al. Two PHOSPHATE-TRANSPORTER1 genes in cotton enhance tolerance to phosphorus starvation [J]. Plant Physiol Biochem, 2023, 204: 108128. |
| 30 | Qiao KK, Lv JY, Chen LL, et al. GhSTP18, a member of sugar transport proteins family, negatively regulates salt stress in cotton [J]. Physiol Plant, 2023, 175(4): e13982. |
| 31 | Chen BZ, Wang XY, Lv JY, et al. GhN/AINV13 positively regulates cotton stress tolerance by interacting with the 14-3-3 protein [J]. Genomics, 2021, 113(1 Pt 1): 44-56. |
| 32 | Zhang Q, Ma C, Wang X, et al. Genome-wide identification of the light-harvesting chlorophyll a/b binding (Lhc) family in Gossypium hirsutum reveals the influence of GhLhcb2.3 on chlorophyll a synthesis [J]. Plant Biol, 2021, 23(5): 831-842. |
| 33 | Hano C, Drouet S, Lainé E. Virus-induced gene silencing (VIGS) in flax (Linum usitatissimum L.) seed coat: description of an effective procedure using the transparent testa 2 gene as a selectable marker [J]. Methods Mol Biol, 2020, 2172: 233-242. |
| [1] | 刘洁, 王飞, 陶婷, 张玉静, 陈浩婷, 张瑞星, 石玉, 张毅. 过表达SlWRKY41提高番茄幼苗抗旱性[J]. 生物技术通报, 2025, 41(2): 107-118. |
| [2] | 黄颖, 遇文婧, 刘雪峰, 刁桂萍. 山新杨谷胱甘肽转移酶基因的生物信息学与表达模式分析[J]. 生物技术通报, 2025, 41(2): 248-256. |
| [3] | 丁若羲, 豆硕, 安叶芝, 孔文慧, 郭文静, 张冬梅, 王省芬, 马峙英, 吴立柱. VIGS技术在棉花全生长周期中的应用拓展研究[J]. 生物技术通报, 2025, 41(2): 58-64. |
| [4] | 孔青洋, 张晓龙, 李娜, 张晨洁, 张雪云, 于超, 张启翔, 罗乐. 单叶蔷薇GRAS转录因子家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 210-220. |
| [5] | 张曼玉, 董嘉诚, 苟福凡, 弓朝晖, 刘倩, 孙文良, 孔臻, 郝捷, 王敏, 田朝光. 嗜热毁丝霉果胶酯酶MtCE12-1的克隆表达及其酶学性质和应用研究[J]. 生物技术通报, 2024, 40(9): 291-300. |
| [6] | 王茜, 周家燕, 王倩, 邓玉萍, 张敏慧, 陈静, 杨军, 邹建. 向日葵YABBY基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(8): 199-211. |
| [7] | 韩凯, 周永顺, 张凯月, 王路, 高剑峰, 陈福龙. 三株小球藻抗旱性能评价[J]. 生物技术通报, 2024, 40(8): 244-254. |
| [8] | 杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141. |
| [9] | 侯文婷, 孙琳, 张艳军, 董合忠. 基因编辑技术在棉花种质创新和遗传改良中的应用[J]. 生物技术通报, 2024, 40(7): 68-77. |
| [10] | 胡雅丹, 伍国强, 刘晨, 魏明. MYB转录因子在调控植物响应逆境胁迫中的作用[J]. 生物技术通报, 2024, 40(6): 5-22. |
| [11] | 秦健, 李振月, 何浪, 李俊玲, 张昊, 杜荣. 肌源性细胞分化的单细胞转录谱变化及细胞间通讯分析[J]. 生物技术通报, 2024, 40(6): 330-342. |
| [12] | 文洁, 杜元欣, 吴安波, 杨广容, 鲁敏, 安华明, 南红. 刺梨SOD基因家族鉴定与表达模式分析[J]. 生物技术通报, 2024, 40(5): 153-166. |
| [13] | 陈春林, 李白雪, 李金玲, 杜清洁, 李猛, 肖怀娟. 甜瓜CmEPF基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(4): 130-138. |
| [14] | 陈强, 黄馨慧, 张峥, 张冲, 柳叶飞. 褪黑素对薄皮甜瓜采后软化和乙烯合成的影响[J]. 生物技术通报, 2024, 40(4): 139-147. |
| [15] | 张玉, 石磊, 巩檑, 聂峰杰, 杨江伟, 刘璇, 杨文静, 张国辉, 颉瑞霞, 张丽. 马铃薯WOX基因家族的鉴定及在离体再生和非生物胁迫中的表达分析[J]. 生物技术通报, 2024, 40(3): 170-180. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||