








生物技术通报 ›› 2024, Vol. 40 ›› Issue (5): 153-166.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1138
文洁( ), 杜元欣, 吴安波, 杨广容, 鲁敏, 安华明, 南红(
), 杜元欣, 吴安波, 杨广容, 鲁敏, 安华明, 南红( )
)
收稿日期:2023-12-02
出版日期:2024-05-26
发布日期:2024-06-13
通讯作者:
南红,女,博士,讲师,研究方向:果树分子生物学、比较基因组学和大数据分析;E-mail: hnan@gzu.edu.cn作者简介:文洁,女,硕士研究生,研究方向:果树分子生物学;E-mail: 18286604835@163.com
基金资助:
WEN Jie( ), DU Yuan-xin, WU An-bo, YANG Guang-rong, LU Min, AN Hua-ming, NAN Hong(
), DU Yuan-xin, WU An-bo, YANG Guang-rong, LU Min, AN Hua-ming, NAN Hong( )
)
Received:2023-12-02
Published:2024-05-26
Online:2024-06-13
摘要:
【目的】超氧化物歧化酶(SOD)是植物体内氧自由基的天然清除剂,对于保护植物免受环境胁迫起着关键作用,了解其在刺梨响应干旱胁迫中的作用,为深入研究刺梨SOD基因家族的功能提供基础。【方法】通过生物信息学对刺梨SOD基因家族进行系统鉴定和分析,对其理化性质、染色体位置、基因结构、亚家族进化、顺式作用元件和WGCNA进行全面分析,并利用RT-qPCR分析刺梨SOD基因家族在干旱胁迫下的表达模式。【结果】刺梨SOD基因家族有9个成员,包括4个Cu/ZnSOD基因、3个MnSOD基因和2个FeSOD基因,分布在5条染色体上。基因结构显示,同一亚家族成员的基序组成较为相似,但内含子/外显子排列和数量差异较大。亚家族进化分析发现,MnSOD亚家族较原始,其在所有蛋白质位置都表现出高度保守性,其次是FeSOD和Cu/ZnSOD,Cu/ZnSOD亚家族各序列之间差异较大,又可以分为3个亚类。顺式作用元件分析显示,该家族基因参与多种植物激素、生长发育以及胁迫响应,特别是RrCSD2启动子区含有类黄酮生物合成元件(MBSI)。进一步通过转录组和WGCNA分析发现,RrCSD2所在模块的基因显著富集到黄酮和黄酮醇生物合成、类黄酮生物合成和花生四烯酸代谢等途径,表明RrCSD2可能参与类黄酮代谢过程。RT-qPCR结果显示,在干旱胁迫下,除RrMSD1和RrMSD2的表达水平显著下降外,其余7个基因的表达水平总体呈现上升趋势,特别是RrCSD2和RrCSD3的表达水平显著上调。【结论】刺梨SOD基因在干旱胁迫中起到重要作用。
文洁, 杜元欣, 吴安波, 杨广容, 鲁敏, 安华明, 南红. 刺梨SOD基因家族鉴定与表达模式分析[J]. 生物技术通报, 2024, 40(5): 153-166.
WEN Jie, DU Yuan-xin, WU An-bo, YANG Guang-rong, LU Min, AN Hua-ming, NAN Hong. Identification and Expression Pattern Analysis of Rosa roxburghii SOD Gene Family[J]. Biotechnology Bulletin, 2024, 40(5): 153-166.
| 基因名称Gene name | 基因ID Gene ID | 上游引物Upstream primer(5'-3') | 下游引物Downstream primer(5'-3') |
|---|---|---|---|
| RrCSD1 | evm.model.Contig149.101 | GATGGCCCAACTACTGTGACT | ACCGTTTGTTGTGTCACCAAG |
| RrCSD2 | evm.model.Contig172.106 | CTGTATTGCCAGAGCTACTGAC | TGCACCCAGAATCCTAACAAC |
| RrCSD3 | evm.model.Contig255.128 | CCGGTGATTTGGGTAACGTCACT | GTTCATGTCCACCTGCCAGTCGTC |
| RrCSD4 | evm.model.Contig428.424 | ACCCAAGAAGATGACGGTCCT | TCGTGTCACCAAACTCATGCAAG |
| RrMSD1 | evm.model.Contig308.148 | ATCATGGACCTCCATCACTT | GTGGAGGAGTCAACTGTATC |
| RrMSD2 | evm.model.Contig308.149 | GATACGGAAGGAGGAGGTGA | GAACCTTGCAAAGAAGCACC |
| RrMSD3 | evm.model.Contig317.61 | GCACCACCAGGCTTACATCACC | CGCTCTGCAACTTAACGACGG |
| RrFSD1 | evm.model.Contig217.78 | TGAAGCCTCCTCCGTATCCA | TATCCACATAACCCCTATGGTG |
| RrFSD2 | evm.model.Contig285.37 | AACCATCTTGTGTCTTGGAA | AACAGGGATATTAGGTTCGC |
| UBQ | - | ATGCAGATTTTGTGAAGAC | ACCACCACGRAGACGGAG |
表1 RT-qPCR引物
Table 1 Primers used for RT-qPCR
| 基因名称Gene name | 基因ID Gene ID | 上游引物Upstream primer(5'-3') | 下游引物Downstream primer(5'-3') |
|---|---|---|---|
| RrCSD1 | evm.model.Contig149.101 | GATGGCCCAACTACTGTGACT | ACCGTTTGTTGTGTCACCAAG |
| RrCSD2 | evm.model.Contig172.106 | CTGTATTGCCAGAGCTACTGAC | TGCACCCAGAATCCTAACAAC |
| RrCSD3 | evm.model.Contig255.128 | CCGGTGATTTGGGTAACGTCACT | GTTCATGTCCACCTGCCAGTCGTC |
| RrCSD4 | evm.model.Contig428.424 | ACCCAAGAAGATGACGGTCCT | TCGTGTCACCAAACTCATGCAAG |
| RrMSD1 | evm.model.Contig308.148 | ATCATGGACCTCCATCACTT | GTGGAGGAGTCAACTGTATC |
| RrMSD2 | evm.model.Contig308.149 | GATACGGAAGGAGGAGGTGA | GAACCTTGCAAAGAAGCACC |
| RrMSD3 | evm.model.Contig317.61 | GCACCACCAGGCTTACATCACC | CGCTCTGCAACTTAACGACGG |
| RrFSD1 | evm.model.Contig217.78 | TGAAGCCTCCTCCGTATCCA | TATCCACATAACCCCTATGGTG |
| RrFSD2 | evm.model.Contig285.37 | AACCATCTTGTGTCTTGGAA | AACAGGGATATTAGGTTCGC |
| UBQ | - | ATGCAGATTTTGTGAAGAC | ACCACCACGRAGACGGAG |
| 基因名称 Gene name | 基因ID Gene ID | 氨基酸 Amino acid/aa | 分子量 Molecular weight/Da | 等电点 pI | 亚细胞定位 Location |
|---|---|---|---|---|---|
| RrCSD1 | evm.model.Contig149.101 | 161 | 16 424.23 | 5.81 | 线粒体Mitochondria |
| RrCSD2 | evm.model.Contig172.106 | 252 | 26 826.36 | 4.82 | 线粒体Mitochondria |
| RrCSD3 | evm.model.Contig255.128 | 126 | 12 912.35 | 5.68 | 过氧化物酶体Peroxysome |
| RrCSD4 | evm.model.Contig428.424 | 224 | 23 278.95 | 5.79 | 叶绿体Chloroplast |
| RrMSD1 | evm.model.Contig308.148 | 196 | 21 612.59 | 7.73 | 线粒体Mitochondria |
| RrMSD2 | evm.model.Contig308.149 | 235 | 26 494.16 | 6.16 | 线粒体Mitochondria |
| RrMSD3 | evm.model.Contig317.61 | 180 | 19 243.86 | 7.07 | 线粒体Mitochondria |
| RrFSD1 | evm.model.Contig217.78 | 268 | 30 684.68 | 5.43 | 线粒体Mitochondria |
| RrFSD2 | evm.model.Contig285.37 | 270 | 31 123.38 | 6.33 | 线粒体Mitochondria |
表2 RrSOD基因理化性质
Table 2 Physical and chemical properties of RrSOD genes
| 基因名称 Gene name | 基因ID Gene ID | 氨基酸 Amino acid/aa | 分子量 Molecular weight/Da | 等电点 pI | 亚细胞定位 Location |
|---|---|---|---|---|---|
| RrCSD1 | evm.model.Contig149.101 | 161 | 16 424.23 | 5.81 | 线粒体Mitochondria |
| RrCSD2 | evm.model.Contig172.106 | 252 | 26 826.36 | 4.82 | 线粒体Mitochondria |
| RrCSD3 | evm.model.Contig255.128 | 126 | 12 912.35 | 5.68 | 过氧化物酶体Peroxysome |
| RrCSD4 | evm.model.Contig428.424 | 224 | 23 278.95 | 5.79 | 叶绿体Chloroplast |
| RrMSD1 | evm.model.Contig308.148 | 196 | 21 612.59 | 7.73 | 线粒体Mitochondria |
| RrMSD2 | evm.model.Contig308.149 | 235 | 26 494.16 | 6.16 | 线粒体Mitochondria |
| RrMSD3 | evm.model.Contig317.61 | 180 | 19 243.86 | 7.07 | 线粒体Mitochondria |
| RrFSD1 | evm.model.Contig217.78 | 268 | 30 684.68 | 5.43 | 线粒体Mitochondria |
| RrFSD2 | evm.model.Contig285.37 | 270 | 31 123.38 | 6.33 | 线粒体Mitochondria |
| 序号No. | 基因ID Gene ID | 物种Species | 基因名称Gene name | 分类Classifying |
|---|---|---|---|---|
| 1 | FvH4_6g04231.t2 | 野草莓F. vesca | FvSOD1 | Cu/ZnSOD |
| 2 | FvH4_3g30620.t1 | FvSOD2 | Cu/ZnSOD | |
| 3 | FvH4_1g28280.t1 | FvSOD3 | Cu/ZnSOD | |
| 4 | FvH4_5g01090.t1 | FvSOD4 | Cu/ZnSOD | |
| 5 | FvH4_7g25730.t1 | FvSOD5 | MnSOD | |
| 6 | FvH4_4g37110.t1 | FvSOD6 | FeSOD | |
| 7 | FvH4_7g26501.t1 | FvSOD7 | FeSOD | |
| 8 | RchiOBHm_Chr3g0452901 | 月季R. chinensis | RcSOD1 | Cu/ZnSOD |
| 9 | RchiOBHm_Chr5g0056701 | RcSOD2 | Cu/ZnSOD | |
| 10 | RchiOBHm_Chr3g0496871 | RcSOD3 | Cu/ZnSOD | |
| 11 | RchiOBHm_Chr2g0126391 | RcSOD4 | Cu/ZnSOD | |
| 12 | RchiOBHm_Chr1g0371971 | RcSOD5 | MnSOD | |
| 13 | RchiOBHm_Chr4g0446901 | RcSOD6 | FeSOD | |
| 14 | RchiOBHm_Chr5g0072741 | RcSOD7 | MnSOD | |
| 15 | RchiOBHm_Chr1g0373101 | RcSOD8 | FeSOD | |
| 16 | RchiOBHm_Chr1g0318061 | RcSOD9 | FeSOD |
表3 野草莓与月季SOD基因家族成员
Table 3 SOD gene family members of F. vesca and R. chinensis
| 序号No. | 基因ID Gene ID | 物种Species | 基因名称Gene name | 分类Classifying |
|---|---|---|---|---|
| 1 | FvH4_6g04231.t2 | 野草莓F. vesca | FvSOD1 | Cu/ZnSOD |
| 2 | FvH4_3g30620.t1 | FvSOD2 | Cu/ZnSOD | |
| 3 | FvH4_1g28280.t1 | FvSOD3 | Cu/ZnSOD | |
| 4 | FvH4_5g01090.t1 | FvSOD4 | Cu/ZnSOD | |
| 5 | FvH4_7g25730.t1 | FvSOD5 | MnSOD | |
| 6 | FvH4_4g37110.t1 | FvSOD6 | FeSOD | |
| 7 | FvH4_7g26501.t1 | FvSOD7 | FeSOD | |
| 8 | RchiOBHm_Chr3g0452901 | 月季R. chinensis | RcSOD1 | Cu/ZnSOD |
| 9 | RchiOBHm_Chr5g0056701 | RcSOD2 | Cu/ZnSOD | |
| 10 | RchiOBHm_Chr3g0496871 | RcSOD3 | Cu/ZnSOD | |
| 11 | RchiOBHm_Chr2g0126391 | RcSOD4 | Cu/ZnSOD | |
| 12 | RchiOBHm_Chr1g0371971 | RcSOD5 | MnSOD | |
| 13 | RchiOBHm_Chr4g0446901 | RcSOD6 | FeSOD | |
| 14 | RchiOBHm_Chr5g0072741 | RcSOD7 | MnSOD | |
| 15 | RchiOBHm_Chr1g0373101 | RcSOD8 | FeSOD | |
| 16 | RchiOBHm_Chr1g0318061 | RcSOD9 | FeSOD |
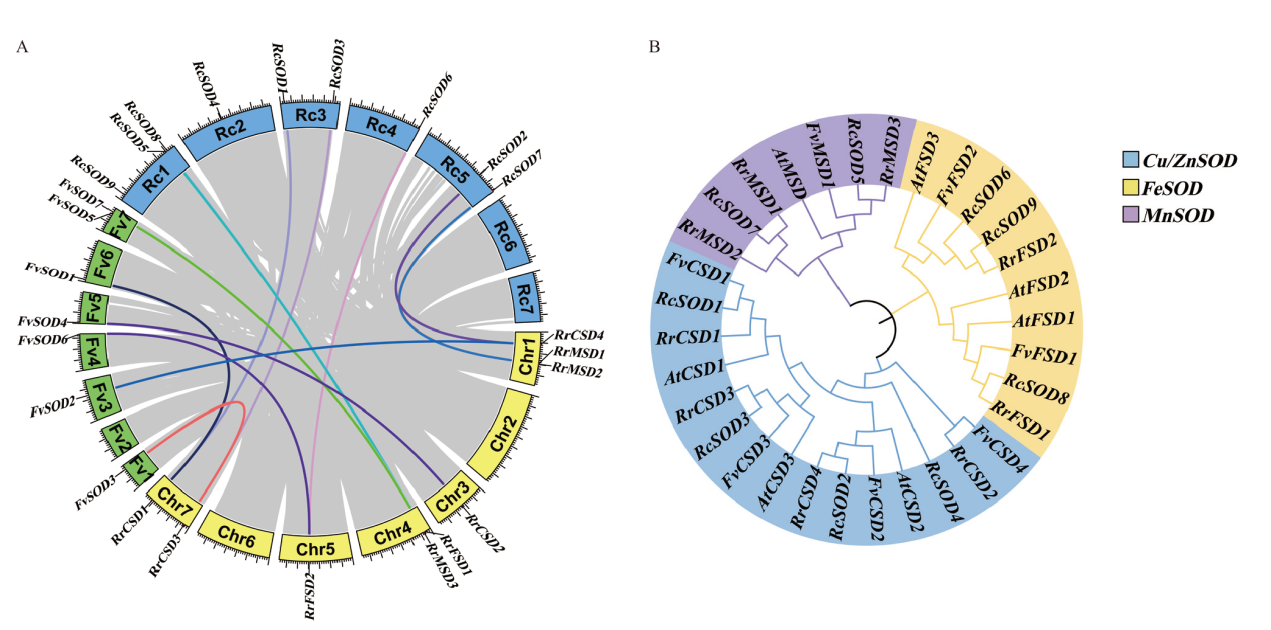
图2 SOD基因家族系统进化分析 A:刺梨、月季和野草莓SOD基因家族共线性分析;B:4个物种的SOD基因家族系统进化分析。At:拟南芥,Rr:刺梨,Fv:野草莓,Rc:月季。利用MEGA X软件构建系统进化树(邻接法),步数设置为1 000
Fig. 2 Phylogenetic analysis of SOD gene family A: Colinear analysis of SOD gene family in R. roxburghii, R. chinensis, and F. vesca. B: Phylogenetic analysis of SOD gene family in four species. At: A. thaliana, Rr: R. roxburghii, Fv: F. vesca, Rc: R. chinensis. The phylogenetic tree was construct using MEGA X with neighbor-joining method, and bootstrap was set to 1 000
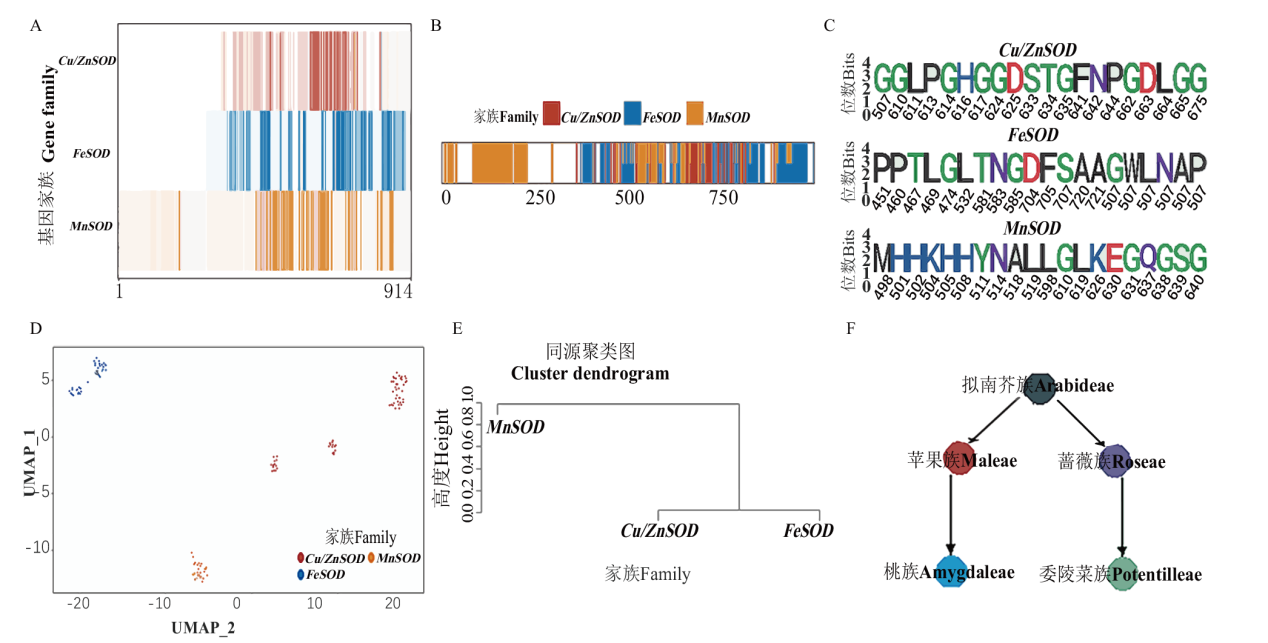
图5 蔷薇科SOD基因亚家族进化分析 A:SOD亚家族保守位点分布热图;B:SOD亚家族特异位点分布图;C:SOD亚家族20个最保守特异位点Weblog图;D:SOD亚家族UMAP聚类图;E:SOD亚家族同源聚类图;F:蔷薇科基因谱系关系
Fig. 5 Analysis of SOD gene subfamily evolution in Rosaceae A: Heatmap showing the conservation of each site in three SOD subfamilies. B: Distribution of the specific sites in three SOD subfamilies. C: Weblog of the 20 most conserved specific site of each SOD subfamily. D: UMAP clustering diagram of SOD subfamilies. E: SOD subfamily homology order. F: Gene lineage relationships of Rosaceae
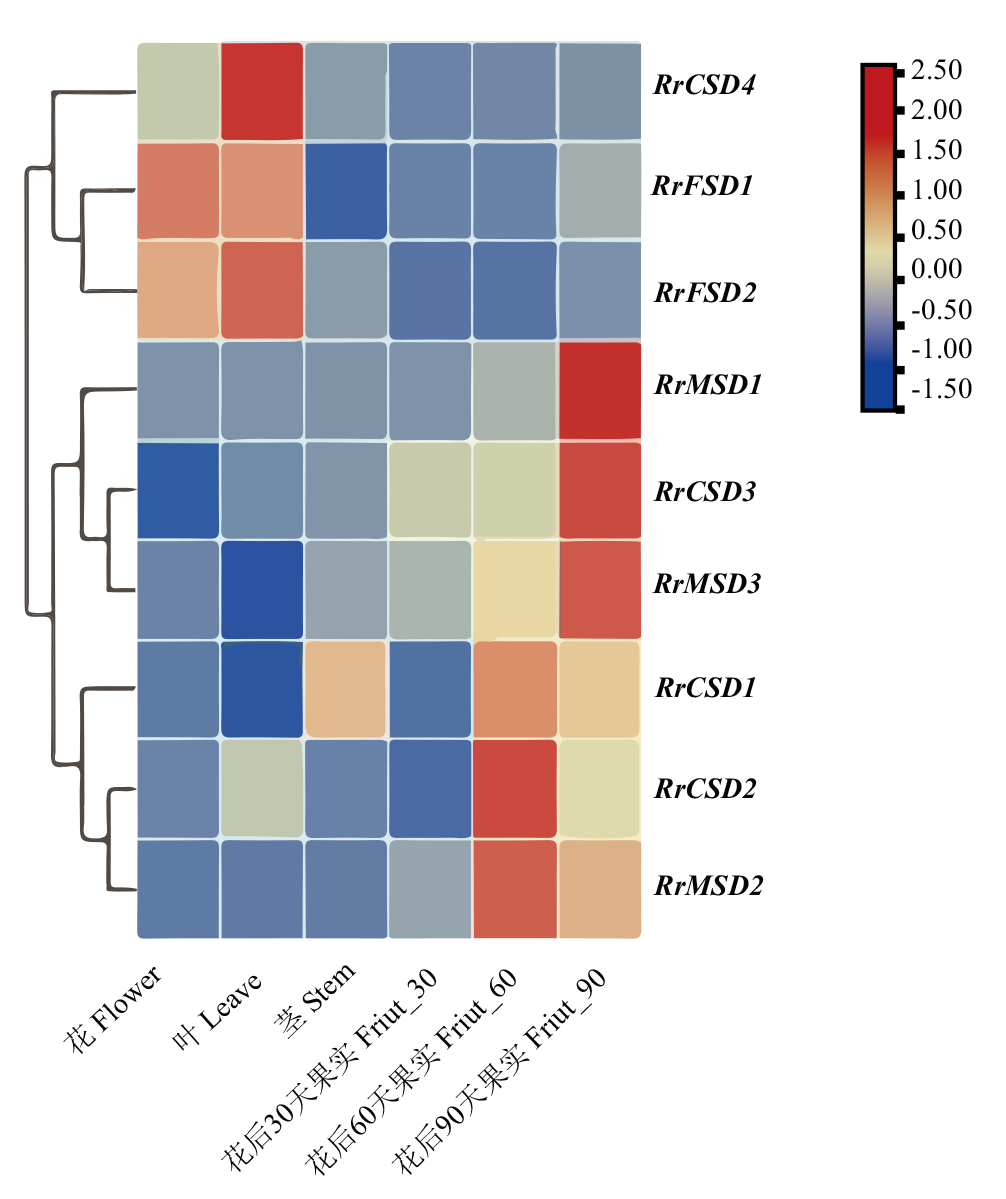
图8 刺梨花、叶、茎和果实不同发育阶段RrSOD基因表达
Fig. 8 Expressions of RrSOD genes in the flowers, leaves, stems and fruits at different developmental stages of R. roxburghii

图9 RrSOD基因WGCNA分析 A:共表达模块与层次聚类树;B:KEGG富集分析
Fig. 9 WGCNA analysis of RrSOD genes A: Co-expression module and hierarchical clustering tree. B: KEGG enrichment analysis
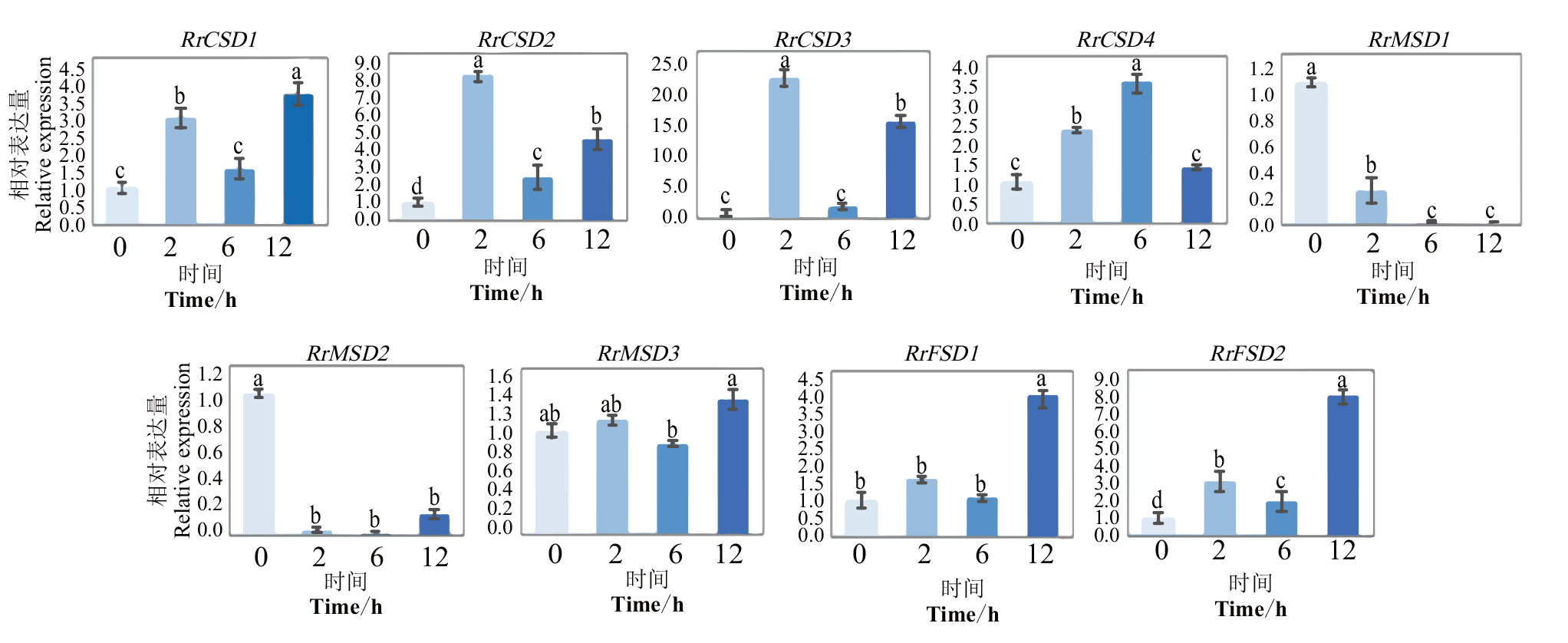
图10 干旱胁迫下RrSOD基因的相对表达分析 不同小写字母表示在0.05水平差异显著
Fig. 10 Relative expression analysis of RrSOD genes under drought stress Different lowercase letters indicate significant difference at 0.05 level
| [1] | 陈仪坤, 杨倩, 何睿. 我国刺梨产业发展存在的问题及对策研究[J]. 中国市场, 2021,(8): 49-50. |
| Chen YK, Yang Q, He R. Research on the problems and countermeasures of Rosa roxburghii industry development[J]. China Market, 2021,(8): 49-50. | |
| [2] | 王朋强, 鲁敏, 覃家雪, 等. 贵州省刺梨产业发展分析与展望--基于毕节市龙凤村的调研[J]. 农业展望, 2023, 19(9): 66-72. |
| Wang PQ, Lu M, Qin JX, et al. Analysis and prospect of Rosa Roxburghii Tratt Industry development in Guizhou province-based on the investigation of Longfeng village in Bijie city[J]. Agricultural Outlook, 2023, 19(9): 66-72. | |
| [3] | 何嵩涛, 刘国琴, 刘进平, 等. 刺梨对土壤干旱胁迫的生理响应[J]. 中国农业科学, 2002,(10): 1243-1248. |
| He ST, Liu GQ, Liu JP, et al. Physiological response to soil drought stress in Rosa roxburghii[J]. Scientia Agricultura Sinica, 2002,(10): 1243-1248. | |
| [4] | 代稳, 王金凤, 杨洪. 喀斯特地区水资源合理配置总控结构研究[J]. 宁夏工程技术, 2017, 16(2): 188-192. |
| Dai W, Wng JF, Yang H. The conceptual framework for rational allocation of water resources in Karst region[J]. Ningxia Engineering Technology, 2017, 16(2): 188-192. | |
| [5] | 李小永, 周玮, 赵晴, 等. 黔中喀斯特水源涵养区不同植被类型土壤水分特征研究[J]. 黑龙江农业科学, 2023,(8): 87-93. |
| Li XY, Zhou W, Zhao Q, et al. Study on soil moisture characteristics under different vegetation types of karst water conservation area in the middle region of Guizhou province[J]. Heilongjiang Agricultural Sciences, 2023,(8): 87-93. | |
| [6] |
Kar RK. Plant responses to water stress: Role of reactive oxygen species[J]. Plant signal behav, 2011, 6(11): 1741-1745.
doi: 10.4161/psb.6.11.17729 pmid: 22057331 |
| [7] | Gill SS, Anjum NA, Gill R, et al. Superoxide dismutase-mentor of abiotic stress tolerance in crop plants[J]. Environmental Science and Pollution Research, 2015, 22(14): 10375-10394. |
| [8] | Abreu IA, Cabelli DE. Superoxide dismutases-a review of the metal-associated mechanistic variations[J]. Biochimica et Biophysica Acta - Proteins and Proteomics, 2010, 1804(2): 263-274. |
| [9] | Jiang W, Yang L, He Y, et al. Genome-wide identification and transcriptional expression analysis of superoxide dismutase(SOD)family in wheat(Triticum aestivum)[J]. PeerJ, 2019, 7(2): e8062. |
| [10] |
Gangwar R, Kumari P, Chatrath A, et al. Characterisation of recombinant thermostable manganese-superoxide dismutase(NeMnSOD)from Nerium oleander[J]. Molecular Biology Reports, 2020, 47(5): 3251-3270.
doi: 10.1007/s11033-020-05374-x pmid: 32297289 |
| [11] | Wang W, Xia MX, Chen J, et al. Gene expression characteristics and regulation mechanisms of superoxide dismutase and its physiological roles in plants under stress[J]. Biochemistry, 2016, 81(5): 465-480. |
| [12] | Zhang X, Zhang LT, Chen YY, et al. Genome-wide identification of the SOD gene family and expression analysis under drought and salt stress in barley[J]. Plant Growth Regulation, 2021, 94(1): 49-60. |
| [13] | Yu WC, Kong GH, Chao JQ, et al. Genome-wide identification of the rubber tree superoxide dismutase(SOD)gene family and analysis of its expression under abiotic stress[J]. Peer J, 2022, 10(20): e14251. |
| [14] | Negi NP, Shrivastava DC, Sharma V, et al. Overexpression of CuZnSOD from Arachis hypogaea alleviates salinity and drought stress in tobacco[J]. Plant Cell Reports, 2015, 34(7): 1109-1126. |
| [15] | Zhang Q, Ruan J, Mumm R, et al. Dynamic changes in the an-tioxidative defense system in the tea plant reveal the photoprotection-mediated temporal accumulation of flavonoids under full sunlight exposure[J]. Plant and Cell Physiology, 2022, 63(11): 1695-1708. |
| [16] | Wang M, Zhang Y, Zhu C, et al. EkFLS overexpression promotes flavonoid accumulation and abiotic stress tolerance in plant[J]. Physiologia Plantarum, 2021, 172(4): 1966-1982. |
| [17] |
孙明慧, 吴琼, 刘丹丹, 等. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1408 |
| Sun MH, Wu Q, Liu DD, et al. Cloning and expression analysis of CsTMFs gene in tea plant[J]. Biotechnology Bulletin, 2023, 39(7): 151-159. | |
| [18] | Lu M, An H, Li L. Genome survey sequencing for the characterization of the genetic background of Rosa roxburghii Tratt and leaf ascorbate metabolism genes[J]. PLoS One, 2016, 11(2): e0147530. |
| [19] | Potter SC, Luciani A, Eddy SR, et al. HMMER web server: 2018 update[J]. Nucleic Acids Research, 2018, 46(W1): W200-W204. |
| [20] | Letunic L, Khedkar S, Bork P. SMART: recent updates, new developments and status in 2020[J]. Nucleic Acids Research, 2020, 49(D1): D458-D460. |
| [21] |
Gasteiger E, Gattiker A, Hoogland C, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis[J]. Nucleic Acids Research, 2003, 31(13): 3784-3788.
doi: 10.1093/nar/gkg563 pmid: 12824418 |
| [22] |
Chen C, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for Interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [23] |
Hu B, Jin J, Guo A, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296-1297.
doi: 10.1093/bioinformatics/btu817 pmid: 25504850 |
| [24] | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Research, 2009, 37: W202-W208. |
| [25] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Research, 2002, 30(1): 325-327. |
| [26] |
Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 2018, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [27] | Letunic I, Bork P. Interactive tree of life(iTOL)v3: an online tool for the display and annotation of phylogenetic and other trees[J]. Nucleic Acids Research, 2016, 44(W1): W242-W245. |
| [28] |
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability[J]. Molecular Biology and Evolution, 2013, 30(4): 772-780.
doi: 10.1093/molbev/mst010 pmid: 23329690 |
| [29] | Zhang Z, Zhu MM, Xie Q, et al. CProtMEDIAS: clustering of amino acid sequences encoded by gene families by MErging and DIgitizing Aligned Sequences[J]. Briefings in Bioinformatics, 2022, 23(4): bbac276. |
| [30] | Huang X, Yan H, Zhai L, et al. Characterization of the Rosa roxburghii Tratt transcriptome and analysis of MYB genes[J]. Plos One, 2019, 14(3): e0203014. |
| [31] | Lu M, Ma WT, Liu YQ, et al. Transcriptome analysis reveals candidate lignin-related genes and transcription factors in Rosa roxburghii during fruit ripening[J]. Plant Molecular Biology Reporter, 2020, 38(2): 331-342. |
| [32] | Chen S, Zhou Y, Chen Y, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): i884-i890. |
| [33] |
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements[J]. Nature Methods, 2015, 12(4): 357-360.
doi: 10.1038/nmeth.3317 pmid: 25751142 |
| [34] | Yu GC, Wang LG, Han YY, et al. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. OMICS: A Journal of Integrative Biology, 2012, 16(5): 284-287. |
| [35] | Rychlik W. OLIGO 7 primer analysis software[J]. Methods in Molecular Biology(Clifton, NJ), 2007, 402: 35-60. |
| [36] | Ye J, Coulouris G, Zaretskaya I, et al. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction[J]. BMC Bioinformatics, 2012, 13(1): 134. |
| [37] | Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications[J]. BMC Bioinformatics, 2009, 10(1): 421. |
| [38] | 赵熳秋, 南红, 鲁敏, 等. 刺梨CaM基因的鉴定及其在果实中的表达特点[J]. 农业生物技术学报, 2022, 30(09): 1724-1736. |
| Zhao MQ, Nan H, Lu M, et al. Identification of CaM genes and its expression characteristics in fruit of Rosa roxburghii[J]. Journal of Agricultural Biotechnology, 2022, 30(09): 1724-1736. | |
| [39] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [40] | Li G, Hu F, Zhang Y, et al. Comparative genomic analysis of superoxide dismutase(SOD)genes in three Rosaceae species and expression analysis in Pyrus bretschneideri[J]. Physiology and Molecular Biology of Plants, 2021, 27(1): 39-52. |
| [41] | Corpas FJ, Fernandez-Ocana A, Carreras A, et al. The expression of different superoxide dismutase forms is cell-type dependent in olive(Olea europaea L.) leaves[J]. Plant Cell Physiology, 2006, 47(7): 984-994. |
| [42] |
Wang W, Zhang X, Pei, Deng FN, et al. Genome-wide characterization and expression analyses of superoxide dismutase(SOD)genes in Gossypium hirsutum[J]. BMC Genomics, 2017, 18(1): 376.
doi: 10.1186/s12864-017-3768-5 pmid: 28499417 |
| [43] | 魏婧, 徐畅, 李可欣, 等. 超氧化物歧化酶的研究进展与植物抗逆性[J]. 植物生理学报, 2020, 56(12): 2571-2584. |
| Wei J, Xu C, Li KX, et al. Progress on superoxide dismutase and plant stress resistance[J]. Plant Physiology Journal, 2020, 56(12): 2571-2584. | |
| [44] | Carrier MC, Ng Kwan Lim E, Jeannotte G, et al. Trans-Acting effectors versus RNA Cis-Elements: A tightly knit regulatory mesh[J]. Frontiers in Microbiology, 2020, 11: 609327. |
| [45] | Xu J, Duan X, Yang J, et al. Coupled expression of Cu/Zn-superoxide dismutase and catalase in cassava improves tolerance against cold and drought stresses[J]. Plant Signaling Behavior, 2013, 8(6): e24525. |
| [46] | Huang X, Sun G, Li Q, et al. Transcriptome analysis reveals regulatory networks and hub genes in the flavonoid metabolism of Rosa roxburghii[J]. Horticulturae, 2023, 9(2): 233. |
| [47] | Nakabayashi R, Yonekura-Sakakibara K, Urano K, et al. Enhancement of oxidative and drought tolerance in arabidopsis by overaccumulation of antioxidant flavonoids[J]. Plant Journal, 2014, 77(3): 367-379. |
| [48] | Baozhu L, Ruonan F, Yanting F, et al. The flavonoid biosynthesis regulator PFG3 confers drought stress tolerance in plants by promoting flavonoid accumulation[J]. Environmental and Experimental Botany, 2022, 196: 104792. |
| [49] | Huang H, Jiao YX, Tong Y, et al. Comparative analysis of drought-responsive biochemical and transcriptomic mechanisms in two Dendrobium officinale genotypes[J]. Industrial Crops and Products, 2023, 199: 116766. |
| [50] | Shen SS, Yan WP, Xie S, et al. Physiological and transcriptional analysis reveals the response mechanism of Camellia vietnamensis Huang to drought stress[J]. International Journal of Molecular Sciences, 2022, 23(19): 11801. |
| [1] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [2] | 刘雯锦, 马瑞, 刘升燕, 杨江伟, 张宁, 司怀军. 马铃薯StCIPK11的克隆及响应干旱胁迫分析[J]. 生物技术通报, 2023, 39(9): 147-155. |
| [3] | 丁凯鑫, 王立春, 田国奎, 王海艳, 李凤云, 潘阳, 庞泽, 单莹. 烯效唑缓解植物干旱损伤的研究进展[J]. 生物技术通报, 2023, 39(6): 1-11. |
| [4] | 王春语, 李政君, 王平, 张丽霞. 高粱表皮蜡质缺失突变体sb1抗旱生理生化分析[J]. 生物技术通报, 2023, 39(5): 160-167. |
| [5] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [6] | 王琪, 胡哲, 富薇, 李光哲, 郝林. 伯克霍尔德氏菌GD17对黄瓜幼苗耐干旱的调节[J]. 生物技术通报, 2023, 39(3): 163-175. |
| [7] | 于波, 秦晓惠, 赵杨. 植物感应干旱信号的机制[J]. 生物技术通报, 2023, 39(11): 6-17. |
| [8] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [9] | 冯策婷, 江律, 刘鑫颖, 罗乐, 潘会堂, 张启翔, 于超. 单叶蔷薇NAC基因家族鉴定及干旱胁迫响应分析[J]. 生物技术通报, 2023, 39(11): 283-296. |
| [10] | 鄢梦雨, 韦晓薇, 曹婧, 兰海燕. 异子蓬SabHLH169基因的克隆及抗旱功能分析[J]. 生物技术通报, 2023, 39(11): 328-339. |
| [11] | 关志秀, 汪燕, 梁成刚, 韦春玉, 黄娟, 陈庆富. 苦荞FtCBL基因的鉴定及对干旱与高钙胁迫的响应[J]. 生物技术通报, 2022, 38(8): 101-109. |
| [12] | 陈佳敏, 刘永杰, 马锦绣, 李丹, 公杰, 赵昌平, 耿洪伟, 高世庆. 小麦组蛋白甲基化酶在杂交种中干旱胁迫表达模式分析[J]. 生物技术通报, 2022, 38(7): 51-61. |
| [13] | 于国红, 刘朋程, 李磊, 李明哲, 崔海英, 郝洪波, 郭安强. 不同基因型马铃薯对干旱胁迫的生理响应[J]. 生物技术通报, 2022, 38(5): 56-63. |
| [14] | 董亚茹, 赵东晓, 耿兵, 李云芝, 王照红. 桑树MnERF2的表达分析[J]. 生物技术通报, 2022, 38(11): 112-121. |
| [15] | 张彤彤, 郑登俞, 吴忠义, 张中保, 于荣. 玉米NF-Y转录因子基因ZmNF-YB13响应干旱和盐胁迫的功能分析[J]. 生物技术通报, 2022, 38(10): 115-123. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||