








生物技术通报 ›› 2025, Vol. 41 ›› Issue (5): 175-185.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0997
• 研究报告 • 上一篇
吴娅1,2( ), 姚润1, 杨含婷1,2, 刘微1,2, 杨帅2,3, 宋驰2(
), 姚润1, 杨含婷1,2, 刘微1,2, 杨帅2,3, 宋驰2( ), 陈士林2(
), 陈士林2( )
)
收稿日期:2024-10-12
出版日期:2025-05-26
发布日期:2025-06-05
通讯作者:
宋驰,男,教授,研究方向 :药用物种基因组及资源鉴定;E-mail: songchi@cdutcm.edu.cn作者简介:吴娅,女,硕士研究生,研究方向 :中药资源与鉴定;E-mail: wuyaya1880@126.com
基金资助:
WU Ya1,2( ), YAO Run1, YANG Han-ting1,2, LIU Wei1,2, YANG Shuai2,3, SONG Chi2(
), YAO Run1, YANG Han-ting1,2, LIU Wei1,2, YANG Shuai2,3, SONG Chi2( ), CHEN Shi-lin2(
), CHEN Shi-lin2( )
)
Received:2024-10-12
Published:2025-05-26
Online:2025-06-05
摘要:
目的 短链脱氢酶/还原酶(SDR)属于NADPH依赖性氧化还原酶超家族,是植物代谢研究中重要的基因家族。对凤梨薄荷(Mentha suaveolens ‘Variegata’)中SDR基因家族进行全基因组鉴定和分析,为解析SDR基因功能与薄荷醇生物合成研究提供理论依据。 方法 基于凤梨薄荷单倍型基因组数据,利用生物信息学方法鉴定凤梨薄荷SDR基因家族,并进行理化特性、保守基序、染色体定位等分析,结合转录联合代谢组数据将SDR基因与薄荷醇合成途径上重要基因及化合物进行共表达分析,以及RT-qPCR分析与单萜类化合物强烈正相关的MsSDR基因表达模式。 结果 共鉴定出142个MsSDRs基因不均匀分布在12条染色体上,编码232-765个氨基酸。系统发育分析将MsSDRs蛋白分为5种类型:经典型(C)、延伸型(E)、非经典型(A)、发散型(D)以及未知型(U)。此外,MsSDRs的启动子序列分析识别出与光响应和胁迫反应相关的顺式作用元件。基因表达图谱显示超过一半MsSDRs至少在一个组织中表达。共表达分析发现32个SDR基因与单萜类化合物呈现较强相关性,RT-qPCR结果与RNA-Seq数据趋势基本一致。 结论 对凤梨薄荷SDR基因家族的全面分析发现142个MsSDRs基因参与了凤梨薄荷生长发育的多个阶段,多个MsSDRs基因与薄荷醇生物合成途径上的单萜类化合物相关,推测其可能参与薄荷醇的生物合成。
吴娅, 姚润, 杨含婷, 刘微, 杨帅, 宋驰, 陈士林. 凤梨薄荷SDR基因家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(5): 175-185.
WU Ya, YAO Run, YANG Han-ting, LIU Wei, YANG Shuai, SONG Chi, CHEN Shi-lin. Genome-wide Identification and Expression Analysis of SDR Gene Family in Mentha suaveolens ‘Variegata’[J]. Biotechnology Bulletin, 2025, 41(5): 175-185.
基因1 Gene 1 | 基因1 Gene 2 | 非同义替换率 Ka | 同义替换率 Ks | Ka/Ks |
|---|---|---|---|---|
| MsSDR1E1 | MsSDR1E3 | 0.08 | 1.16 | 0.07 |
| MsSDR2E2 | MsSDR2E3 | 0.07 | 1.30 | 0.05 |
| MsSDR31E3 | MsSDR31E2 | 0.17 | 0.66 | 0.25 |
| MsSDR50E4 | MsSDR50E5 | 0.23 | 2.21 | 0.10 |
| MsSDR50E2 | MsSDR50E7 | 0.07 | 0.46 | 0.16 |
| MsSDR67E1 | MsSDR67E2 | 0.05 | 0.78 | 0.06 |
| MsSDR93E1 | MsSDR93E2 | 0.03 | 1.69 | 0.02 |
| MsSDR117E1 | MsSDR117E3 | 0.28 | 1.70 | 0.17 |
| MsSDR367E1 | MsSDR367E2 | 0.11 | 1.22 | 0.09 |
| MsSDR132C6 | MsSDR132C1 | 0.29 | 3.11 | 0.09 |
| MsSDR152C2 | MsSDR152C1 | 0.13 | 0.89 | 0.15 |
| MsSDR119C3 | MsSDR119C2 | 0.11 | 2.87 | 0.04 |
| MsSDR114C10 | MsSDR114C12 | 0.19 | 1.99 | 0.10 |
| MsSDR110C15 | MsSDR110C8 | 0.42 | 0.73 | 0.58 |
表1 MsSDR 基因 Ka/Ks 分析
Table 1 Ka/Ks analysis of MsSDR genes
基因1 Gene 1 | 基因1 Gene 2 | 非同义替换率 Ka | 同义替换率 Ks | Ka/Ks |
|---|---|---|---|---|
| MsSDR1E1 | MsSDR1E3 | 0.08 | 1.16 | 0.07 |
| MsSDR2E2 | MsSDR2E3 | 0.07 | 1.30 | 0.05 |
| MsSDR31E3 | MsSDR31E2 | 0.17 | 0.66 | 0.25 |
| MsSDR50E4 | MsSDR50E5 | 0.23 | 2.21 | 0.10 |
| MsSDR50E2 | MsSDR50E7 | 0.07 | 0.46 | 0.16 |
| MsSDR67E1 | MsSDR67E2 | 0.05 | 0.78 | 0.06 |
| MsSDR93E1 | MsSDR93E2 | 0.03 | 1.69 | 0.02 |
| MsSDR117E1 | MsSDR117E3 | 0.28 | 1.70 | 0.17 |
| MsSDR367E1 | MsSDR367E2 | 0.11 | 1.22 | 0.09 |
| MsSDR132C6 | MsSDR132C1 | 0.29 | 3.11 | 0.09 |
| MsSDR152C2 | MsSDR152C1 | 0.13 | 0.89 | 0.15 |
| MsSDR119C3 | MsSDR119C2 | 0.11 | 2.87 | 0.04 |
| MsSDR114C10 | MsSDR114C12 | 0.19 | 1.99 | 0.10 |
| MsSDR110C15 | MsSDR110C8 | 0.42 | 0.73 | 0.58 |
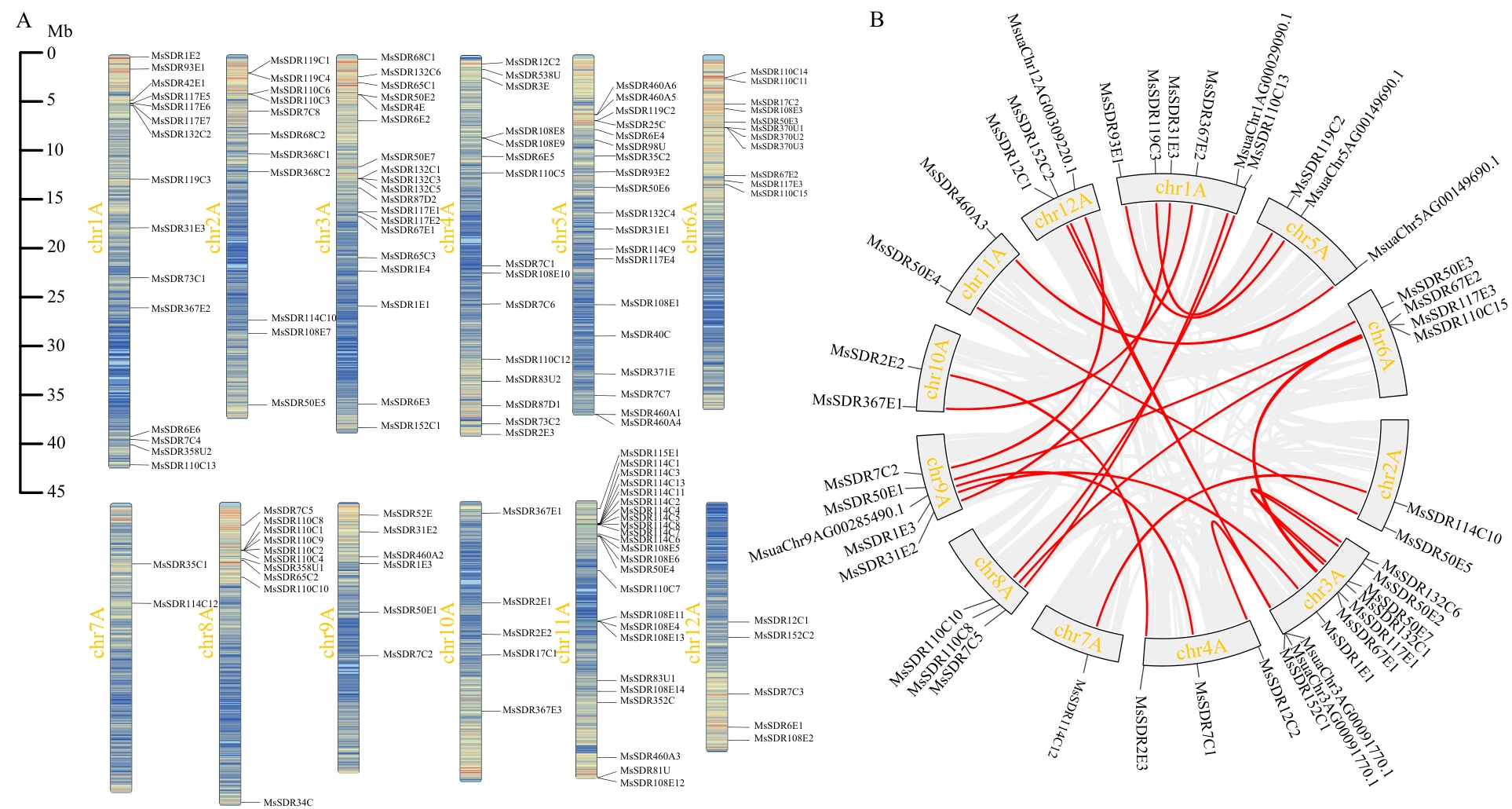
图3 凤梨薄荷MsSDR基因的染色体定位以及共线性分析A:染色体定位;B:共线性分析
Fig. 3 Chromosomal localization of the MsSDRs in M. suaveolens and analysis of collinearityA: Localization of chromosomes. B: Colinearity analysis

图4 凤梨薄荷MsSDR基因的KEGG通路富集分析(A),GO富集分析(B)以及顺式作用元件预测统计(C)
Fig. 4 KEGG pathway enrichment analysis (A), GO enrichment analysis(B), and cis-acting element prediction statistics (C) for the MsSDRs gene in M. suaveolens

图5 凤梨薄荷MsSDR基因的不同组织的表达分析A:凤梨薄荷SDR基因在不同组织的表达模式;B:共表达网络图,绿色代表正相关,黄色代表负相关;C:6个MsSDR基因在不同组织中的表达谱,图中误差线表示标准偏差
Fig. 5 Expression analysis of MsSDRs in different tissues of M. suaveolensA: Expression patterns of SDR genes in different tissues of M. suaveolens. B: Co-expression network diagram, with green representing positive correlation and yellow representing negative correlation. C: Expression profiles of six MsSDR genes in various tissues. The error line in the figure refers to the standard deviation
| 1 | 郭明遗, 邓艳, 杜前红, 等. 青李薄荷酒发酵工艺优化及风味成分分析 [J]. 食品与机械, 2023, 39(6): 201-209. |
| Guo MY, Deng Y, Du QH, et al. Fermentation process optimization and flavor composition analysis of plum peppermint wine [J]. Food Mach, 2023, 39(6): 201-209. | |
| 2 | 温亚娟, 项丽玲, 苗明三. 薄荷的现代应用研究 [J]. 中医学报, 2016, 31(12): 1963-1965. |
| Wen YJ, Xiang LL, Miao MS. Modern applied research on Mentha haplocalyx [J]. Acta Chin Med, 2016, 31(12): 1963-1965. | |
| 3 | 陈军. 我国薄荷属植物特性及利用(Ⅱ) [J]. 林产工业, 2011, 48(4): 54-55. |
| Chen J. Characteristics and utilization of Mentha in China (Ⅱ) [J]. China For Prod Ind, 2011, 48(4): 54-55. | |
| 4 | Ramzi A, El Ouali Lalami A, Zoubi YE, et al. Insecticidal effect of wild-grown Mentha pulegium and Rosmarinus officinalis essential oils and their main monoterpenes against Culex pipiens (Diptera: Culicidae) [J]. Plants, 2022, 11(9): 1193. |
| 5 | 尹东阁, 王开心, 刘曼婷, 等. 《中华人民共和国药典》2020年版收载含冰片、薄荷的中药成方制剂质量标准分析 [J]. 中华中医药学刊, 2023, 41(2): 24-31. |
| Yin DG, Wang KX, Liu MT, et al. Analysis of quality standard of Chinese medicine preparations containing Bingpian(borneol)and Bohe(mint)in 2020 edition of Chinese pharmacopoeia [J]. Chin Arch Tradit Chin Med, 2023, 41(2): 24-31. | |
| 6 | 侴桂新. 薄荷属植物化学分类学研究及其意义 [J]. 中草药, 1991, 22(11): 519-525. |
| Chou GX. Studies on the chemical taxonomy of Mentha spp. and its significance [J]. Chinese Traditional and Herbal Drugs, 1991, 22(11): 519-525. | |
| 7 | 杨翠云, 安馨, 万晶琼, 等. 不同化学型薄荷挥发油抗菌和抗氧化活性研究 [J]. 食品科技, 2021, 46(1): 185-192. |
| Yang CY, An X, Wan JQ, et al. Antibacterial and antioxidant activities of different chemotypes of essential oils from Mentha haplocalyx briq [J]. Food Sci Technol, 2021, 46(1): 185-192. | |
| 8 | Moreno L, Bello R, Primo-Yúfera E, et al. Pharmacological properties of the methanol extract from Mentha suaveolens Ehrh [J]. Phytother Res, 2002, 16(): S10-S13. |
| 9 | Angiolella L, Vavala E, Sivric S, et al. Letter: in vitro activity of Mentha Suaveolens essential oil against cryptococcus neoformans and dermatophytes [J]. International Journal of Essential Oil Therapeutics, 2010, 4: 35-36. |
| 10 | Ferreira A, Proença C, Serralheiro MM, et al. The in vitro screening for acetylcholinesterase inhibition and antioxidant activity of medicinal plants from Portugal [J]. J Ethnopharmacol, 2006, 108(1): 31-37. |
| 11 | Lahlou S, Ferreira Lima Carneiro-Leão R, Leal-Cardoso JH. Cardiovascular effects of the essential oil of Mentha x Villosa in DOCA-salt-hypertensive rats [J]. Phytomedicine, 2002, 9(8): 715-720. |
| 12 | Kimbaris AC, González-Coloma A, Andrés MF, et al. Biocidal compounds from Mentha sp. essential oils and their structure-activity relationships [J]. Chem Biodivers, 2017, 14(3). DOI:10.1002/cbdv.201600270 . |
| 13 | Yang HT, Wang YF, Liu W, et al. Genome-wide pan-GPCR cell libraries accelerate drug discovery [J]. Acta Pharm Sin B, 2024, 14(10): 4296-4311. |
| 14 | Moummou H, Kallberg Y, Tonfack LB, et al. The plant short-chain dehydrogenase (SDR) superfamily: genome-wide inventory and diversification patterns [J]. BMC Plant Biol, 2012, 12: 219. |
| 15 | Lin XJ, Huang LX, Liang HL, et al. Genome-wide identification and functional characterization of borneol dehydrogenases in Wurfbainia villosa [J]. Planta, 2023, 258(4): 69. |
| 16 | Agarwal P, Pathak S, Kumar RS, et al. Short-chain dehydrogenase/reductase, PsDeHase, from opium poppy: putative involvement in papaverine biosynthesis [J]. Plant Cell Tissue Organ Cult, 2020, 143(2): 431-440. |
| 17 | Rahier A, Bergdoll M, Génot G, et al. Homology modeling and site-directed mutagenesis reveal catalytic key amino acids of 3beta-hydroxysteroid-dehydrogenase/C4-decarboxylase from Arabidopsis [J]. Plant Physiol, 2009, 149(4): 1872-1886. |
| 18 | Yu SH, Sun QG, Wu JX, et al. Genome-wide identification and characterization of short-chain dehydrogenase/reductase (SDR) gene family in Medicago truncatula [J]. Int J Mol Sci, 2021, 22(17): 9498. |
| 19 | Hwang SG, Lin NC, Hsiao YY, et al. The Arabidopsis short-chain dehydrogenase/reductase 3, an abscisic acid deficient 2 homolog, is involved in plant defense responses but not in ABA biosynthesis [J]. Plant Physiol Biochem, 2012, 51: 63-73. |
| 20 | Sato-Masumoto N, Ito M. Isolation and characterization of isopiperitenol dehydrogenase from piperitenone-type Perilla [J]. Biol Pharm Bull, 2014, 37(5): 847-852. |
| 21 | Su XJ, Yang LL, Wang DL, et al. 1 K medicinal plant genome database: an integrated database combining genomes and metabolites of medicinal plants [J]. Hortic Res, 2022, 9: uhac075. |
| 22 | Yang HT, Wang C, Zhou GR, et al. A haplotype-resolved gap-free genome assembly provides novel insight into monoterpenoid diversification in Mentha suaveolens 'Variegata' [J]. Hortic Res, 2024, 11(3): uhae022. |
| 23 | Leng L, Xu ZC, Hong BX, et al. Cepharanthine analogs mining and genomes of Stephania accelerate anti-coronavirus drug discovery [J]. Nat Commun, 2024, 15(1): 1537. |
| 24 | Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11 [J]. Mol Biol Evol, 2021, 38(7): 3022-3027. |
| 25 | Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments [J]. Nucleic Acids Res, 2019, 47(W1): W256-W259. |
| 26 | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| 27 | Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks [J]. Genome Res, 2003, 13(11): 2498-2504. |
| 28 | Bharati R, Sen MK, Kumar R, et al. Systematic identification of suitable reference genes for quantitative real-time PCR analysis in Melissa officinalis L [J]. Plants, 2023, 12(3): 470. |
| 29 | Kram BW, Xu WW, Carter CJ. Uncovering the Arabidopsis thaliana nectary transcriptome: investigation of differential gene expression in floral nectariferous tissues [J]. BMC Plant Biol, 2009, 9: 92. |
| 30 | Cannon SB, Mitra A, Baumgarten A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana [J]. BMC Plant Biol, 2004, 4: 10. |
| 31 | An X, Wan JQ, Jiang H, et al. Transcriptome analysis of transcription factors and enzymes involved in monoterpenoid biosynthesis in different chemotypes of Mentha haplocalyx Briq [J]. PeerJ, 2023, 11: e14914. |
| 32 | Ringer KL, Davis EM, Croteau R. Monoterpene metabolism. Cloning, expression, and characterization of (-)-isopiperitenol/(-)-carveol dehydrogenase of peppermint and spearmint [J]. Plant Physiol, 2005, 137(3): 863-872. |
| 33 | Kavanagh KL, Jörnvall H, Persson B, et al. Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes [J]. Cell Mol Life Sci, 2008, 65(24): 3895-3906. |
| 34 | 王帅乐, 肖珂雨, 王维东, 等. 苹果短链脱氢酶基因MdSDR响应腐烂病菌侵染的功能研究 [J]. 核农学报, 2022, 36(11): 2158-2165. |
| Wang SL, Xiao KY, Wang WD, et al. Function exploration of apple short-chain dehydrogenases/reductase in response to Valsa Mali [J]. J Nucl Agric Sci, 2022, 36(11): 2158-2165. | |
| 35 | Malik I, Almrani HA. Volatile oil yield and its components of menthol and limonene of peppermint affected by foliar spray and lighting intensity [J]. IOP Conf Ser: Earth Environ Sci, 2023, 1262(5): 052008. |
| 36 | Zheng YM, Zhu YS, Mao XH, et al. SDR7-6, a short-chain alcohol dehydrogenase/reductase family protein, regulates light-dependent cell death and defence responses in rice [J]. Mol Plant Pathol, 2022, 23(1): 78-91. |
| 37 | Turner GW, Davis EM, Croteau RB. Immunocytochemical localization of short-chain family reductases involved in menthol biosynthesis in peppermint [J]. Planta, 2012, 235(6): 1185-1195. |
| 38 | 李玉, 贾浩田, 耿晓云, 等. 梅SDR基因家族的鉴定及在花开放阶段的表达分析 [J]. 基因组学与应用生物学, 2024, 43(6): 997-1009. |
| Li Y, Jia HT, Geng XY, et al. Identification of SDR gene family in Prunus mume and its expression analysis during flower opening stage [J]. Genom Appl Biol, 2024, 43(6): 997-1009. | |
| 39 | 刘俊, 寇杰锋, 练从龙, 等. 杜仲Trihelix基因家族全基因组鉴定及表达模式分析 [J]. 中国中药杂志, 2024, 49(22): 6093-6106. |
| Liu J, Kou JF, Lian CL, et al. Genome-wide identification and expression pattern analysis of Eucommia ulmoides Trihelix gene family [J]. China J Chin Mater Med, 2024, 49(22): 6093-6106. | |
| 40 | Qin SS, Wei F, Liang Y, et al. Genome-wide analysis of the R2R3-MYB gene family in Spatholobus suberectus and identification of its function in flavonoid biosynthesis [J]. Front Plant Sci, 2023, 14: 1219019. |
| 41 | Wang X, Liang YF, Shu J, et al. Transcription factor StWRKY1 is involved in monoterpene biosynthesis induced by light intensity in Schizonepeta tenuifolia Briq [J]. Plant Physiol Biochem, 2024, 214: 108871. |
| 42 | Liao BS, Shen XF, Xiang L, et al. Allele-aware chromosome-level genome assembly of Artemisia annua reveals the correlation between ADS expansion and artemisinin yield [J]. Mol Plant, 2022, 15(8): 1310-1328. |
| 43 | Xu J, Liao BS, Yuan L, et al. 50th anniversary of artemisinin: From the discovery to allele-aware genome assembly of Artemisia annua [J]. Mol Plant, 2022, 15(8): 1243-1246. |
| 44 | Wang Y, Zhang H, Ri HC, et al. Deletion and tandem duplications of biosynthetic genes drive the diversity of triterpenoids in Aralia elata [J]. Nat Commun, 2022, 13(1): 2224. |
| 45 | Chen HY, Guo MX, Dong ST, et al. A chromosome-scale genome assembly of Artemisia argyi reveals unbiased subgenome evolution and key contributions of gene duplication to volatile terpenoid diversity [J]. Plant Commun, 2023, 4(3): 100516. |
| 46 | 房海灵, 李维林, 任冰如, 等. 薄荷属植物的化学成分及药理学研究进展 [J]. 中国药业, 2010, 19(10): 13-17. |
| Fang HL, Li WL, Ren BR, et al. Research advance on chemical constituents and pharmacological activities of Mentha L [J]. China Pharm, 2010, 19(10): 13-17. |
| [1] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| [2] | 秦健, 李振月, 何浪, 李俊玲, 张昊, 杜荣. 肌源性细胞分化的单细胞转录谱变化及细胞间通讯分析[J]. 生物技术通报, 2024, 40(6): 330-342. |
| [3] | 刘换换, 杨立春, 李火根. 北美鹅掌楸LtMYB305基因的克隆及功能分析[J]. 生物技术通报, 2024, 40(4): 179-188. |
| [4] | 胡明月, 杨宇, 郭仰东, 张喜春. 低温胁迫下番茄SlMYB96的功能分析[J]. 生物技术通报, 2023, 39(4): 236-245. |
| [5] | 陈楚雯, 李洁, 赵瑞鹏, 刘媛, 吴锦波, 李志雄. 藏鸡GPX3基因的克隆、组织表达谱研究及功能预测[J]. 生物技术通报, 2023, 39(3): 311-320. |
| [6] | 张玉娟, 黎冬华, 宫慧慧, 崔新晓, 高春华, 张秀荣, 游均, 赵军胜. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析[J]. 生物技术通报, 2023, 39(11): 308-317. |
| [7] | 陈浩婷, 张玉静, 刘洁, 代泽敏, 刘伟, 石玉, 张毅, 李天来. 低磷胁迫下番茄转录因子WRKY6功能分析[J]. 生物技术通报, 2023, 39(10): 136-147. |
| [8] | 于秋琳, 马婧怡, 赵盼, 孙鹏芳, 何玉美, 刘世彪, 郭惠红. 绞股蓝GpMIR156a和GpMIR166b的克隆与功能分析[J]. 生物技术通报, 2022, 38(7): 186-193. |
| [9] | 党瑗, 李维, 苗向, 修宇, 林善枝. 山杏油体蛋白基因PsOLE4克隆及其调控油脂累积功能分析[J]. 生物技术通报, 2022, 38(11): 151-161. |
| [10] | 杨宏亮, 袁桢, 钱徐佳志, 徐大伟. 大麦Thionin-like基因家族基因表达谱分析[J]. 生物技术通报, 2022, 38(10): 140-147. |
| [11] | 马亚男, 卢旭, 魏云春, 李康, 魏若男, 李胜, 马绍英. 葡萄AKR基因家族的鉴定和组织特异性表达分析[J]. 生物技术通报, 2021, 37(8): 141-151. |
| [12] | 季文博, 王会, 柴志欣, 王吉坤, 信金伟, 钟金城. 牦牛HYOU1基因克隆及组织表达分析[J]. 生物技术通报, 2019, 35(3): 123-131. |
| [13] | 杨意宏, 徐浩, 陈段芬, 高志民. 高等植物赤霉素3-β-双氧化酶基因研究进展[J]. 生物技术通报, 2018, 34(3): 18-22. |
| [14] | 王娜, 龚娜, 刘国丽, 马晓颖, 杨镇, 杨涛. 内生菌次生代谢产物提高玉米渗透胁迫抗性的表达谱分析[J]. 生物技术通报, 2016, 32(7): 87-92. |
| [15] | 刘妍, 孟志刚, 孙国清, 王远, 周焘, 郭三堆, 张锐. 陆地棉GhPYR1基因的克隆和功能分析[J]. 生物技术通报, 2016, 32(2): 90-99. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||