








生物技术通报 ›› 2025, Vol. 41 ›› Issue (7): 181-192.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0105
李霞( ), 张泽伟, 刘泽军, 王楠, 郭江波, 辛翠花, 张彤, 简磊(
), 张泽伟, 刘泽军, 王楠, 郭江波, 辛翠花, 张彤, 简磊( )
)
收稿日期:2025-01-24
出版日期:2025-07-26
发布日期:2025-07-22
通讯作者:
简磊,女,博士,讲师,研究方向 :作物抗逆基因功能及调控网络;E-mail: jianleijane@imust.edu.cn作者简介:李霞,女,硕士研究生,研究方向 :马铃薯基因功能解析;E-mail: 599570370@qq.com
基金资助:
LI Xia( ), ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei(
), ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei( )
)
Received:2025-01-24
Published:2025-07-26
Online:2025-07-22
摘要:
目的 MYB转录因子通过调控逆境响应基因,在植物抵御非生物胁迫中发挥着重要作用。旨在探究马铃薯转录因子StMYB96在响应非生物胁迫中的功能。 方法 克隆StMYB96基因,对其进行生物信息学分析、亚细胞定位和表达模式分析。通过体外表达StMYB96并结合DNA亲和纯化测序技术(DNA affinity purification sequencing, DAP-seq),在全基因组范围内鉴定StMYB96的靶基因及其参与的非生物胁迫响应途径。 结果 StMYB96的开放阅读框为1 002 bp,编码333个氨基酸。多序列比对结果显示,StMYB96与番茄(Solanum lycopersicum)SlMYB306、辣椒(Capsicum annuum)CaMYB306和枸杞(Lycium barbarum)LbMYB306的亲缘关系较近。亚细胞定位结果表明,StMYB96定位于细胞核中。RT-qPCR分析显示,StMYB96在马铃薯幼苗的根、茎和叶中均有表达,其中,在茎中表达量最高。在模拟干旱胁迫下,StMYB96的表达量显著上调;而在低温处理下,其表达量持续下调。通过DAP-seq技术,鉴定出StMYB96的3个结合位点,在全基因组范围内鉴定到8 837个靶基因,其启动子区域包含StMYB96的结合位点,这些靶基因广泛参与多种代谢途径。其中,类黄酮生物合成途径相关基因和脂肪酸延伸途径相关基因均受StMYB96直接调控。 结论 StMYB96可能通过调控不同的代谢途径参与马铃薯对干旱和低温胁迫的响应。
李霞, 张泽伟, 刘泽军, 王楠, 郭江波, 辛翠花, 张彤, 简磊. 马铃薯转录因子StMYB96的克隆及功能研究[J]. 生物技术通报, 2025, 41(7): 181-192.
LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato[J]. Biotechnology Bulletin, 2025, 41(7): 181-192.
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 用途 Usage |
|---|---|---|
| StMYB96-F | CGACGACAAGACCGTGACCATGGGAAGACCACCTTGCTG | 基因克隆 |
| StMYB96-R | GAGGAGAAGAGCCGTCGAAAAAAGTCAGCAGATTCAC | Gene cloning |
| StMYB96-qRT-F | TCAAAGGGACAGTGGGAGAG | 荧光定量PCR |
| StMYB96-qRT-R | AGAGCTGGATCCTGTAACGG | RT-qPCR |
| StEF1α-qRT-F | GACAAGCGTGTTATTGAGAGG | 内参基因 |
| StEF1α-qRT-R | CACAGTGCAGTAGTACTTAGTG | Reference gene |
| StMYB96-OX-F | CACGGGGGACTCTAGAATGGGAAGACCACCTTGCTG | 过表达载体的构建 |
| StMYB96-OX-R | GGGGAAATTCGAGCTCTCAAAAAAAGTCAGCAGATT | Construction of overexpression vector |
表1 本研究所用的引物
Table 1 Primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 用途 Usage |
|---|---|---|
| StMYB96-F | CGACGACAAGACCGTGACCATGGGAAGACCACCTTGCTG | 基因克隆 |
| StMYB96-R | GAGGAGAAGAGCCGTCGAAAAAAGTCAGCAGATTCAC | Gene cloning |
| StMYB96-qRT-F | TCAAAGGGACAGTGGGAGAG | 荧光定量PCR |
| StMYB96-qRT-R | AGAGCTGGATCCTGTAACGG | RT-qPCR |
| StEF1α-qRT-F | GACAAGCGTGTTATTGAGAGG | 内参基因 |
| StEF1α-qRT-R | CACAGTGCAGTAGTACTTAGTG | Reference gene |
| StMYB96-OX-F | CACGGGGGACTCTAGAATGGGAAGACCACCTTGCTG | 过表达载体的构建 |
| StMYB96-OX-R | GGGGAAATTCGAGCTCTCAAAAAAAGTCAGCAGATT | Construction of overexpression vector |

图1 马铃薯StMYB96的克隆及序列分析A:马铃薯StMYB96基因PCR产物电泳图(M: DNA marker; P: StMYB96 PCR扩增产物);B:StMYB96基因结构;C:StMYB96蛋白结构域分析;D:StMYB96的基因序列及推导的氨基酸序列(绿色和粉色标记部分:MYB结构域;*:终止密码子)
Fig. 1 Cloning and sequence analysis of StMYB96 in S. tuberosum L.A: Electrophoretogram of PCR product of StMYB96 gene in S. tuberosum L. (M: DNA marker; P: StMYB96 PCR amplified product). B: Gene structure of StMYB96. C: Conserved domain analysis ofStMYB96 protein. D: StMYB96 gene sequence and deduced amino acid sequence (Green marked nucleotide sequence and pink marked amino acid sequence indicate MYB domain; the asterisk (*) refers to stop codon)

图2 StMYB96蛋白的二级、三级结构预测A: StMYB96蛋白的二级结构预测(大写字母:StMYB96蛋白氨基酸序列;小写字母:不同的二级结构,c表示无规则卷曲,h表示α-螺旋,e代表示延伸链,t表示β转角); B:StMYB96蛋白的三级结构预测
Fig. 2 Prediction of secondary and tertiary structure of StMYB96 proteinA: The secondary structure prediction of StMYB96 protein. Capital letters indicate amino acid sequence of StMYB96 protein. Lowercase letters indicate different secondary structures, where c, h, e and t indicate random coil, α-helix, extended strand and β-turn, respectively. B: The tertiary structure prediction of StMYB96 protein

图3 StMYB96与其他物种同源蛋白的氨基酸序列比对深蓝色标记的部分代表氨基酸完全保守,粉色标记的部分代表氨基酸部分保守,浅蓝色标记的部分代表氨基酸相似
Fig. 3 Multiple sequence alignment between StMYB96 and its homologous proteins from other speciesDark blue indicates amino acids completely conserved, pink indicates amino acids partially conserved, and light blue indicates amino acids similar

图4 StMYB96蛋白系统发育树StMYB96:马铃薯;SlMYB306:番茄(XP_004236011.1);CaMYB306:辣椒(XP_016563223.1);LbMYB306b:枸杞(XP_060205081.1);NtMYB306:烟草(XP_016452464.1);ArMYB96:猕猴桃(GFZ17560.1);AtMYB96:拟南芥(NP_201053.2);AtMYB94:拟南芥(OAP04445.1)
Fig. 4 Phylogenetic tree of StMYB96 proteinStMYB96: Solanum tuberosum (PGSC0003DMG400019535); SlMYB306: Solanum lycopersicum (XP_004236011.1); CaMYB306: Capsicum annuum (XP_016563223.1); LbMYB306b: Lycium barbarum (XP_060205081.1); NtMYB306: Nicotiana tabacum (XP_016452464.1); ArMYB96: Actinidia rufa (GFZ17560.1); AtMYB96: Arabidopsis thaliana (NP_201053.2); AtMYB94 (OAP04445.1)

图5 马铃薯StMYB96亚细胞定位分析A: 35S::StMYB96::GFP重组载体构建示意图; B: 35S::StMYB96::GFP融合蛋白在烟草叶片中的亚细胞定位(从左到右分别是绿色荧光、叶绿体自发荧光、明场、叠加场;标尺=50 µm)
Fig. 5 Subcellular localization of StMYB96 in S. tuberosum L.A: The structure of 35S::StMYB96::GFP vector. B: Subcellular localization of GFP protein and StMYB96::GFP fusion protein in Nicotiana benthamiana epidermal cells. (From left to right: GFP fluorescence, chloroplast self-luminescence, bright-field, and merged microscope image; Scale bar = 50 µm)

图6 StMYB96在不同组织及非生物胁迫下的表达量分析A: StMYB96在不同组织中的表达量;B: StMYB96在干旱胁迫下的表达量变化;C: StMYB96在低温胁迫下的表达量变化。HT (Hours of treatment)表示处理时间,**表示在P<0.01水平差异显著
Fig. 6 Expression analysis of StMYB96 in various tissues and in response to various abiotic stressesA: Relative expressions of StMYB96 in different tissues. B: Expression profiles of StMYB96 in response to drought stress. C: Expression profiles of StMYB96 in response to low temperature stress. HT: Hours of treatment. ** indicates significant difference at P<0.01
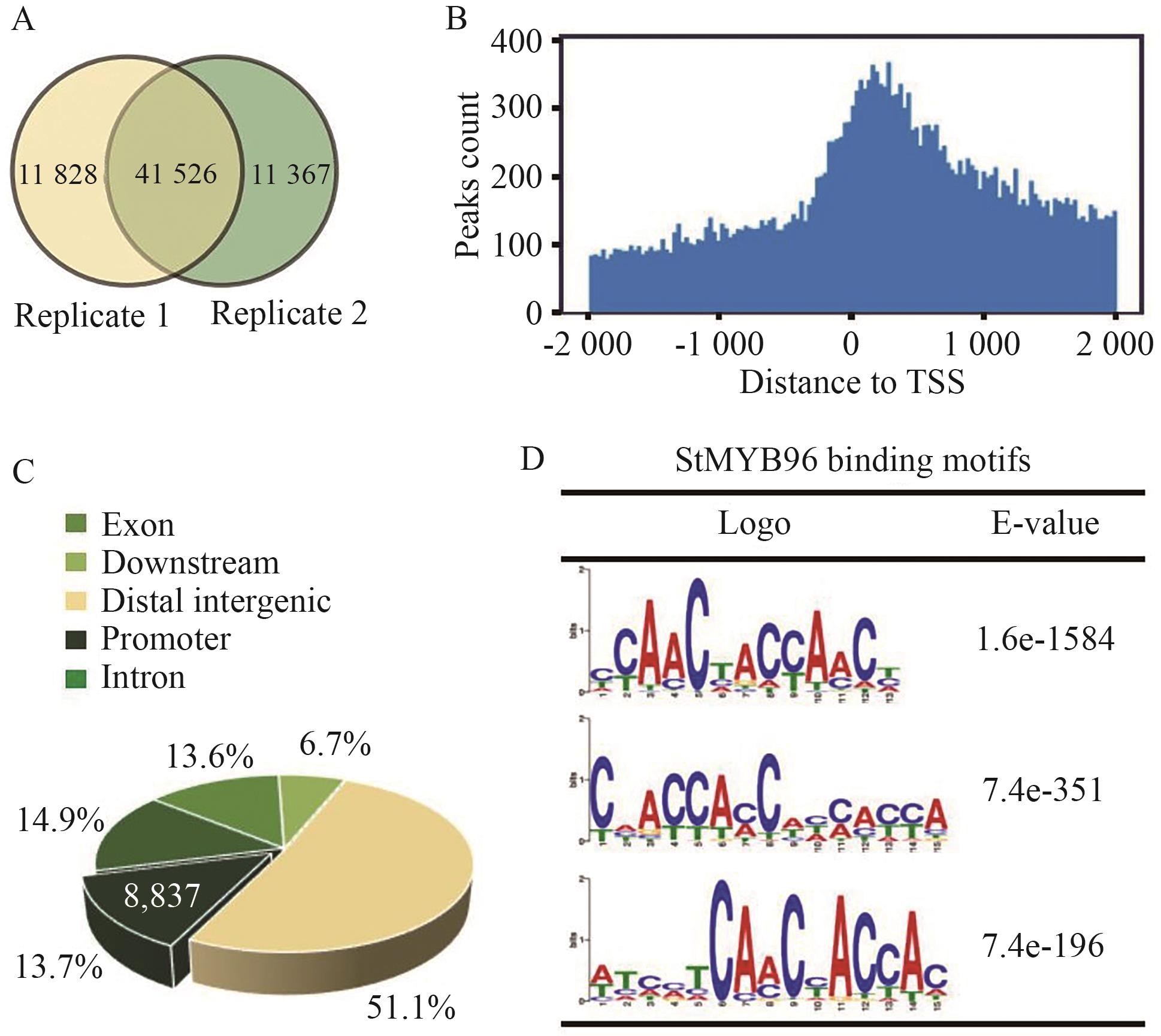
图7 StMYB96直接结合靶点的全基因组分析A: 两个生物学重复中鉴定到的StMYB96与基因组DNA特异性结合的peak数量维恩图;B: StMYB96结合位点在转录起始位点(TSS)两侧-2 000 到 +2 000 bp区域的分布;C: StMYB96结合区域在马铃薯基因组中的分布;D: StMYB96的结合基序
Fig. 7 Genome-wide identification of direct binding sites of StMYB96A: Venn diagram of peak number specifically binding to genomic DNA by StMYB96 identified in two biological replicates. B: Distribution of StMYB96 binding sites related to transcription start sites (TSS ±2 000 bp). C: Distribution of StMYB96 binding sites in potato genome. D: Binding motif of StMYB96

图8 StMYB96靶基因的KEGG通路富集分析和GO功能注释分析A: StMYB96靶基因的KEGG通路富集分析;横坐标为通路对应的Rich factor,纵坐标为通路名称,点的大小表示每个通路下靶基因的数目,点的颜色表示P值的大小;B: StMYB96靶基因的GO功能注释分析;横坐标为GO三个大类(生物过程、分子功能和细胞组分)的下一层级GO term,纵坐标为注释到该term的靶基因数目占比
Fig. 8 KEGG pathway enrichment analysis and GO functional annotation analysis of StMYB96 target genesA: KEGG pathway enrichment analysis of StMYB96 target genes. The x-axis refers to the Rich factor corresponding to the pathways, the y-axis refers to the pathway names, the sizes of the dots indicate the number of target genes in each pathway, and the color of the dot indicates the size of the P-value. B: GO functional annotation analysis of StMYB96 target genes. The x-axis refers to the next-level GO terms under the three main GO categories (biological process, molecular function, and cellular component), and the y-axis refers to the proportion of target genes annotated to each term

图9 StMYB96靶基因中响应干旱和低温胁迫的相关基因A:StMYB96靶基因中参与类黄酮生物合成和脂肪酸延伸途径的基因;B:StMYB96靶基因中CBF信号通路的相关基因;C:StMYB96靶基因中部分功能注释显示与植物抗旱相关的基因。Rep1和Rep2表示两个生物学重复(replicate),Fold enrichment(富集倍数)表示StMYB96在基因启动子区域上的结合富集程度
Fig. 9 StMYB96-target genes involved in the responses to drought and low-temperature stressA: StMYB96-target genes involved in the flavonoid biosynthesis and fatty acid elongation pathway. B: StMYB96-target genes involved in the CBF signaling pathway. C: StMYB96-target genes involved in some functional annotations and drought stress response. Rep1 and Rep2 refer to two biological replicates. Fold enrichment indicates the binding enrichment level of StMYB96 in the promoter regions of genes

图10 StMYB96在马铃薯干旱和低温胁迫响应中的工作模型假说实线代表已知或本研究证实的调控路径,虚线代表假设的调控路径,箭头代表促进作用,T型箭头代表抑制作用
Fig. 10 A working model hypothesis of StMYB96 in response to drought and low-temperature stress in potatoSolid lines indicate known or experimentally validated regulatory pathways, dashed lines indicate hypothesized regulatory pathways, arrows indicate activation, and T-shaped arrows indicate inhibition
| [1] | Wang XP, Niu YL, Zheng Y. Multiple functions of MYB transcription factors in abiotic stress responses [J]. Int J Mol Sci, 2021, 22(11): 6125. |
| [2] | Wu XY, Xia M, Su P, et al. MYB transcription factors in plants: a comprehensive review of their discovery, structure, classification, functional diversity and regulatory mechanism [J]. Int J Biol Macromol, 2024, 282(Pt 2): 136652. |
| [3] | 位欣欣, 兰海燕. 植物MYB转录因子调控次生代谢及逆境响应的研究进展 [J]. 生物技术通报, 2022, 38(8): 12-23. |
| Wei XX, Lan HY. Advances in the regulation of plant MYB transcription factors in secondary metabolism and stress response [J]. Biotechnol Bull, 2022, 38(8): 12-23. | |
| [4] | 胡雅丹, 伍国强, 刘晨, 等. MYB转录因子在调控植物响应逆境胁迫中的作用 [J]. 生物技术通报, 2024, 40(6): 5-22. |
| Hu YD, Wu GQ, Liu C, et al. The role of MYB transcription factor in regulating plant response to adversity stress [J]. China Ind Econ, 2024, 40(6): 5-22. | |
| [5] | Yadav B, Jogawat A, Rahman MS, et al. Secondary metabolites in the drought stress tolerance of crop plants: a review [J]. Gene Rep, 2021, 23: 101040. |
| [6] | Sudiro C, Guglielmi F, Hochart M, et al. A phenomics and metabolomics investigation on the modulation of drought stress by a biostimulant plant extract in tomato (Solanum lycopersicum) [J]. Agronomy, 2022, 12(4): 764. |
| [7] | Qin LM, Sun L, Wei L, et al. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress [J]. Plant J, 2021, 105(4): 1010-1025. |
| [8] | Zhang L, Wang L, Fang YC, et al. Phosphorylated transcription factor PuHB40 mediates ROS-dependent anthocyanin biosynthesis in pear exposed to high light [J]. Plant Cell, 2024, 36(9): 3562-3583. |
| [9] | Naing AH, Kim CK. Abiotic stress-induced anthocyanins in plants: their role in tolerance to abiotic stresses [J]. Physiol Plant, 2021, 172(3): 1711-1723. |
| [10] | Maier A, Schrader A, Kokkelink L, et al. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis [J]. Plant J, 2013, 74(4): 638-651. |
| [11] | Kumar M, Kumar Patel M, Kumar N, et al. Metabolomics and molecular approaches reveal drought stress tolerance in plants [J]. Int J Mol Sci, 2021, 22(17): 9108. |
| [12] | Jiang H, Zhou LJ, Gao HN, et al. The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple [J]. Plant Physiol, 2022, 189(4): 2044-2060. |
| [13] | Wang FB, Kong WL, Wong G, et al. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana [J]. Mol Genet Genomics, 2016, 291(4): 1545-1559. |
| [14] | Chen XS, Wu Y, Yu ZH, et al. BcMYB111 responds to BcCBF2 and induces flavonol biosynthesis to enhance tolerance under cold stress in non-heading Chinese cabbage [J]. Int J Mol Sci, 2023, 24(10): 8670. |
| [15] | Wang FB, Ren XQ, Zhang F, et al. A R2R3-type MYB transcription factor gene from soybean, GmMYB12, is involved in flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis [J]. Plant Biotechnol Rep, 2019, 13(3): 219-233. |
| [16] | Liu TL, Chen TZ, Kan JL, et al. The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants [J]. Plant Biotechnol J, 2022, 20(4): 722-735. |
| [17] | Chen YN, Feng PP, Zhang XW, et al. Silencing of SlMYB50 affects tolerance to drought and salt stress in tomato [J]. Plant Physiol Biochem, 2022, 193: 139-152. |
| [18] | Chen YN, Li L, Tang BY, et al. Silencing of SlMYB55 affects plant flowering and enhances tolerance to drought and salt stress in tomato [J]. Plant Sci, 2022, 316: 111166. |
| [19] | Lee SB, Kim HU, Suh MC. MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis [J]. Plant Cell Physiol, 2016, 57(11): 2300-2311. |
| [20] | Jian L, Kang K, Choi Y, et al. Mutation of OsMYB60 reduces rice resilience to drought stress by attenuating cuticular wax biosynthesis [J]. Plant J, 2022, 112(2): 339-351. |
| [21] | Song Q, Kong LF, Yang XR, et al. PtoMYB142, a poplar R2R3-MYB transcription factor, contributes to drought tolerance by regulating wax biosynthesis [J]. Tree Physiol, 2022, 42(10): 2133-2147. |
| [22] | Cao MX, Liu HZ, Zhang C, et al. Functional analysis of StPHT1;7, a Solanum tuberosum L. phosphate transporter gene, in growth and drought tolerance [J]. Plants, 2020, 9(10): 1384. |
| [23] | Ma X, Yu YN, Jia JH, et al. The pepper MYB transcription factor CaMYB306 accelerates fruit coloration and negatively regulates cold resistance [J]. Sci Hortic, 2022, 295: 110892. |
| [24] | Yu J, Lei B, Zhao HN, et al. Cloning, characterization and functional analysis of NtMYB306a gene reveals its role in wax alkane biosynthesis of tobacco trichomes and stress tolerance [J]. Front Plant Sci, 2022, 13: 1005811. |
| [25] | Yeats TH, Rose JKC. The formation and function of plant cuticles [J]. Plant Physiol, 2013, 163(1): 5-20. |
| [26] | Lian XY, Gao HN, Jiang H, et al. MdKCS2 increased plant drought resistance by regulating wax biosynthesis [J]. Plant Cell Rep, 2021, 40(12): 2357-2368. |
| [27] | Wu JT, Lv SD, Zhao L, et al. Advances in the study of the function and mechanism of the action of flavonoids in plants under environmental stresses [J]. Planta, 2023, 257(6): 108. |
| [28] | 林春草, 陈大伟, 戴均贵. 黄酮类化合物合成生物学研究进展 [J]. 药学学报, 2022, 57(5): 1322-1335. |
| Lin CC, Chen DW, Dai JG. Advances of synthetic biology of flavonoids [J]. Acta Pharm Sin, 2022, 57(5): 1322-1335. | |
| [29] | Liu T, Liu L, Zhou TS, et al. Chalcone isomerase gene (OsCHI3) increases rice drought tolerance by scavenging ROS via flavonoid and ABA metabolic pathways [J]. Crop J, 2025, 13(2): 372-384. |
| [30] | Bao HH, Yuan L, Luo YC, et al. The transcription factor WRKY41-FLAVONOID 3'-HYDROXYLASE module fine-tunes flavonoid metabolism and cold tolerance in potato [J]. Plant Physiol, 2025, 197(3): kiaf070. |
| [31] | Huang L, Hong YB, Zhang HJ, et al. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance [J]. BMC Plant Biol, 2016, 16(1): 203. |
| [32] | Chen YH, Hu L, Punta M, et al. Homologue structure of the SLAC1 anion channel for closing stomata in leaves [J]. Nature, 2010, 467(7319): 1074-1080. |
| [33] | Sun SJ, Qi GN, Gao QF, et al. Protein kinase OsSAPK8 functions as an essential activator of S-type anion channel OsSLAC1, which is nitrate-selective in rice [J]. Planta, 2016, 243(2): 489-500. |
| [34] | Riboni M, Galbiati M, Tonelli C, et al. GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS [J]. Plant Physiol, 2013, 162(3): 1706-1719. |
| [35] | Lim C, Kang K, Shim Y, et al. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways [J]. Plant Physiol, 2022, 188(4): 1900-1916. |
| [36] | Wang F, Chen HW, Li QT, et al. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants [J]. Plant J, 2015, 83(2): 224-236. |
| [37] | Luo X, Bai X, Sun XL, et al. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling [J]. J Exp Bot, 2013, 64(8): 2155-2169. |
| [1] | 张学琼, 潘素君, 李魏, 戴良英. 植物磷酸盐转运蛋白在胁迫响应中的研究进展[J]. 生物技术通报, 2025, 41(7): 28-36. |
| [2] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [3] | 韩燚, 侯昌林, 唐露, 孙璐, 谢晓东, 梁晨, 陈小强. 大麦HvERECTA基因的克隆及功能分析[J]. 生物技术通报, 2025, 41(7): 106-116. |
| [4] | 黄丹丹, 吴云翼, 邹建华, 俞婷, 朱炎辉, 杨梅宏, 董文丽, 高冬丽. 马铃薯StPTST2a基因的克隆及互作分析[J]. 生物技术通报, 2025, 41(7): 172-180. |
| [5] | 龚钰涵, 陈兰, 尚方慧子, 郝灵颖, 刘硕谦. 茶树TRB基因家族鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(7): 214-225. |
| [6] | 魏雨佳, 李岩, 康语涵, 弓晓楠, 杜敏, 涂岚, 石鹏, 于子涵, 孙彦, 张昆. 白颖苔草CrMYB4基因的克隆和表达分析[J]. 生物技术通报, 2025, 41(7): 248-260. |
| [7] | 罗稷林, 栗锦烨, 贾玉鑫. 马铃薯中重力响应调节基因鉴定及功能分析[J]. 生物技术通报, 2025, 41(6): 109-118. |
| [8] | 许慧珍, SHANTWANA Ghimire, RAJU Kharel, 岳云, 司怀军, 唐勋. 马铃薯SUMO E3连接酶基因家族分析及StSIZ1基因的克隆与表达模式分析[J]. 生物技术通报, 2025, 41(6): 119-129. |
| [9] | 段永红, 杨欣, 于冠群, 夏俊俊, 宋陆帅, 白小东, 彭锁堂. 125份马铃薯种质资源遗传多样性及主成分分析[J]. 生物技术通报, 2025, 41(6): 130-143. |
| [10] | 李小欢, 陈相宇, 陶麒宇, 朱玲, 唐铭, 姚银安, 汪丽君. PtoMYB61对毛白杨木质素合成及耐盐性的影响[J]. 生物技术通报, 2025, 41(6): 284-296. |
| [11] | 程慧娟, 王昕, 石小涛, 马东旭, 龚大春, 胡骏鹏, 谢智文. 转录因子CREA敲除对黑曲霉形态和分泌β-葡萄糖苷酶的影响[J]. 生物技术通报, 2025, 41(6): 344-354. |
| [12] | 李锐, 胡婷, 陈树溦, 王尧, 王计平. 紫苏PfMYB80转录因子正向调控花青素的生物合成[J]. 生物技术通报, 2025, 41(6): 243-255. |
| [13] | 郭涛, 艾丽皎, 邹世慧, 周玲, 李学梅. 山茶CjRAV1调控开花延迟的功能研究[J]. 生物技术通报, 2025, 41(6): 208-217. |
| [14] | 程珊, 王会伟, 陈晨, 朱雅婧, 李春鑫, 别海, 王树峰, 陈献功, 张向歌. 油莎豆MYB转录因子基因CeMYB154克隆及耐盐功能分析[J]. 生物技术通报, 2025, 41(6): 218-228. |
| [15] | 黄丹, 彭兵阳, 张盼盼, 焦悦, 吕佳斌. 油茶HD-Zip基因家族鉴定及其在非生物胁迫下的表达分析[J]. 生物技术通报, 2025, 41(6): 191-207. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||