








生物技术通报 ›› 2025, Vol. 41 ›› Issue (6): 218-228.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1182
• 研究报告 • 上一篇
程珊1( ), 王会伟1, 陈晨1, 朱雅婧1, 李春鑫1, 别海2, 王树峰1, 陈献功1, 张向歌1(
), 王会伟1, 陈晨1, 朱雅婧1, 李春鑫1, 别海2, 王树峰1, 陈献功1, 张向歌1( )
)
收稿日期:2024-12-05
出版日期:2025-06-26
发布日期:2025-06-30
通讯作者:
张向歌,男,博士,助理研究员,研究方向 :作物遗传育种;E-mail: maizezxg@163.com作者简介:程珊,女,硕士,研究实习员,研究方向 :作物遗传育种;E-mail: 18790610677@163.com
基金资助:
CHENG Shan1( ), WANG Hui-wei1, CHEN Chen1, ZHU Ya-jing1, LI Chun-xin1, BIE Hai2, WANG Shu-feng1, CHEN Xian-gong1, ZHANG Xiang-ge1(
), WANG Hui-wei1, CHEN Chen1, ZHU Ya-jing1, LI Chun-xin1, BIE Hai2, WANG Shu-feng1, CHEN Xian-gong1, ZHANG Xiang-ge1( )
)
Received:2024-12-05
Published:2025-06-26
Online:2025-06-30
摘要:
目的 油莎豆MYB转录因子基因CeMYB154参与盐胁迫响应过程,克隆及表征油莎豆CeMYB154并分析其耐盐功能,为油莎豆耐盐育种提供分子基础和基因资源。 方法 基于油莎豆参考基因组信息,克隆CeMYB154的全长CDS序列,利用生物信息学软件对其氨基酸序列特征及其启动子上顺式作用元件进行分析,利用分子生物学技术对其进行亚细胞定位和转录激活验证。构建CeMYB154过表达载体,利用农杆菌介导法转化拟南芥,并对过表达株系的耐盐表型和生理指标进行分析。 结果 成功克隆了长度为780 bp的CeMYB154 CDS序列,其编码蛋白序列中2个典型的MYB结构域,属于R2R3型MYB转录因子。蛋白序列比对分析显示,CeMYB154蛋白在多个物种间序列高度保守。启动子区域含有多种激素(如ABA)响应以及MYB响应相关顺式作用元件。此外,亚细胞定位和转录激活分析显示,CeMYB154是一个具有转录激活活性的核转录因子。成功创制了过表达CeMYB154转基因拟南芥,获得了纯合T3转基因阳性株系。在盐胁迫下,与野生型拟南芥相比,转基因植株幼苗生长状态(叶片大小、颜色以及根长)较好;并且,转基因植株中MDA和H2O2含量均显著降低,而CAT、POD和SOD抗氧化酶活性则显著增强,表明过表达CeMYB154基因提高了拟南芥的耐盐性。 结论 油莎豆CeMYB154具有正向调控盐胁迫响应的功能,过表达CeMYB154基因可以增强抗氧化酶活性而提高转基因植株的耐盐性。
程珊, 王会伟, 陈晨, 朱雅婧, 李春鑫, 别海, 王树峰, 陈献功, 张向歌. 油莎豆MYB转录因子基因CeMYB154克隆及耐盐功能分析[J]. 生物技术通报, 2025, 41(6): 218-228.
CHENG Shan, WANG Hui-wei, CHEN Chen, ZHU Ya-jing, LI Chun-xin, BIE Hai, WANG Shu-feng, CHEN Xian-gong, ZHANG Xiang-ge. Cloning of MYB Transcription Factor Gene CeMYB154 and Analysis of Salt Tolerance Function in Cyperus esculentus[J]. Biotechnology Bulletin, 2025, 41(6): 218-228.
| 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) | 用途 Purpose |
|---|---|---|
| MYB154-F | ATGGAGGAAGTGGCATGGAG | 基因克隆 Gene clone |
| MYB154-R | CTAGTATAAAGCAAATTCTGCTGCTTGC | |
| 2300-F | gtacccggggatcctctagagtcgacATGGAGGAAGTGGCATGGAG | 转基因植株检测 Transgenic plant detection |
| 2300-R | ttgctcaccatggtactagtgtcgacGTATAAAGCAAATTCTGCTGCTTGC | |
| GFP-F | ctatttacaattacggatccATGGAGGAAGTGGCATGGAG | 亚细胞定位 Subcellular localization |
| GFP-R | agatcctcctccggagaccTAAAGCAAATTCTGCTGC | |
| BD-F | atggccatggaggccgaattcATGGAGGAAGTGGCATGGAG | 转录激活活性检测 Transcriptional activation activity assays |
| BD-R | tcgacggatccccgggaattcGTATAAAGCAAATTCTGCTGCTTGC | |
| QP-F | GATGGAATTCTGTTGCT | 荧光定量PCR引物 Fluorescent quantitative PCR primer |
| QP-R | CTTTATCTCGTTGTCGG | |
| CetActin-F | ACTCTGGTGATGGTGTGAGC | 油莎豆内参引物 Internal primer of Cyperus esculentus |
| CetActin-R | CCCTCTCTCCGTCAGGATCT | |
| AtActin-F | GGTAACATTGTGCTCAGTGGTGG | 拟南芥内参引物 Internal primer of Arabidopsis thaliana |
| AtActin-R | AACGACCTTAATCTTCATGCTGC |
表1 本研究所用的引物序列
Table 1 Primer sequences used in this study
| 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) | 用途 Purpose |
|---|---|---|
| MYB154-F | ATGGAGGAAGTGGCATGGAG | 基因克隆 Gene clone |
| MYB154-R | CTAGTATAAAGCAAATTCTGCTGCTTGC | |
| 2300-F | gtacccggggatcctctagagtcgacATGGAGGAAGTGGCATGGAG | 转基因植株检测 Transgenic plant detection |
| 2300-R | ttgctcaccatggtactagtgtcgacGTATAAAGCAAATTCTGCTGCTTGC | |
| GFP-F | ctatttacaattacggatccATGGAGGAAGTGGCATGGAG | 亚细胞定位 Subcellular localization |
| GFP-R | agatcctcctccggagaccTAAAGCAAATTCTGCTGC | |
| BD-F | atggccatggaggccgaattcATGGAGGAAGTGGCATGGAG | 转录激活活性检测 Transcriptional activation activity assays |
| BD-R | tcgacggatccccgggaattcGTATAAAGCAAATTCTGCTGCTTGC | |
| QP-F | GATGGAATTCTGTTGCT | 荧光定量PCR引物 Fluorescent quantitative PCR primer |
| QP-R | CTTTATCTCGTTGTCGG | |
| CetActin-F | ACTCTGGTGATGGTGTGAGC | 油莎豆内参引物 Internal primer of Cyperus esculentus |
| CetActin-R | CCCTCTCTCCGTCAGGATCT | |
| AtActin-F | GGTAACATTGTGCTCAGTGGTGG | 拟南芥内参引物 Internal primer of Arabidopsis thaliana |
| AtActin-R | AACGACCTTAATCTTCATGCTGC |

图1 CeMYB154的克隆与生物信息学分析A:CeMYB154基因克隆;B:不同浓度NaCl处理下油莎豆根系中CeMYB154的相对表达水平;C:CeMYB154蛋白结构域分析;D:CeMYB154亚细胞定位预测;E:多物种序列比对;Ce:油莎豆;Ah:花生;Gm:大豆;Zm:玉米;Bna:油菜;At:拟南芥;OS:水稻;St:马铃薯;Ib:甘薯;**表示以WT为对照在P<0.01水平上差异显著,下同
Fig. 1 Cloning and bioinformatics analysis of CeMYB154A: CeMYB154 gene cloning. B: The expressions of CeMYB154 in Cyperus esculentus roots under different concentrations of NaCl treatment. C: CeMYB154 protein domain analysis. D: CeMYB154 subcellular localization prediction. E: Multi-species sequence alignment. Ce: Cyperus esculentus. Ah:: Arachis hypogaea. Gm: Glycine max. Zm: Zea mays. Bna: Brassica napus. At: Arabidopsis thaliana. Os: Oryza sativa. St: Solanum tuberosum. Ib: Ipomoea batatas. ** indicates a significant difference at the P<0.01 level with WT as control, the same below
| 顺式作用元件 Cis-acting element | 序列 Sequence (5′‒3′) | 功能 Function | 数量 Quantity |
|---|---|---|---|
| CAAT-box | CAAT | 启动子和增强子区域共同的顺式作用元件 | 23 |
| TATA-box | TATATA; ATATAT | 转录起始-30附近的核心启动子元件 | 156 |
| MYB-like | TAACCA | MYB识别和结合元件 | 1 |
| MRE | AACCTAA | MYB响应元件 | 1 |
| TCCC-motif | TCTCCCT | 光响应元件 | 1 |
| GT1-motif | GGTTAAT | 光响应元件 | 5 |
| G-box | CACGTC | 光响应元件 | 1 |
| GATA-motif | GATAGGG | 光响应元件 | 1 |
| AAGAA-motif | GAAAGAA | 脱落酸响应元件 | 2 |
| ABRE | ACGTG | 脱落酸响应元件 | 1 |
| TGA-box | TGACGTAA | 生长素响应元件 | 1 |
| TGACG-motif | TGACG | 茉莉酸甲酯响应元件 | 2 |
| ARE | AAACCA | 厌氧诱导调节元件 | 3 |
表2 CeMYB154启动子顺式作用元件分析
Table 2 Analysis of cis-acting elements in CeMYB154 promoter
| 顺式作用元件 Cis-acting element | 序列 Sequence (5′‒3′) | 功能 Function | 数量 Quantity |
|---|---|---|---|
| CAAT-box | CAAT | 启动子和增强子区域共同的顺式作用元件 | 23 |
| TATA-box | TATATA; ATATAT | 转录起始-30附近的核心启动子元件 | 156 |
| MYB-like | TAACCA | MYB识别和结合元件 | 1 |
| MRE | AACCTAA | MYB响应元件 | 1 |
| TCCC-motif | TCTCCCT | 光响应元件 | 1 |
| GT1-motif | GGTTAAT | 光响应元件 | 5 |
| G-box | CACGTC | 光响应元件 | 1 |
| GATA-motif | GATAGGG | 光响应元件 | 1 |
| AAGAA-motif | GAAAGAA | 脱落酸响应元件 | 2 |
| ABRE | ACGTG | 脱落酸响应元件 | 1 |
| TGA-box | TGACGTAA | 生长素响应元件 | 1 |
| TGACG-motif | TGACG | 茉莉酸甲酯响应元件 | 2 |
| ARE | AAACCA | 厌氧诱导调节元件 | 3 |

图2 亚细胞定位EGFP代表绿色荧光;Cy5代表红色荧光;TD代表明场;ALL代表3种光源叠加
Fig. 2 Subcellular localizationEGFP indicates green fluorescence; Cy5 indicates red fluorescence; TD indicates bright field; ALL indicates the superposition of three light sources
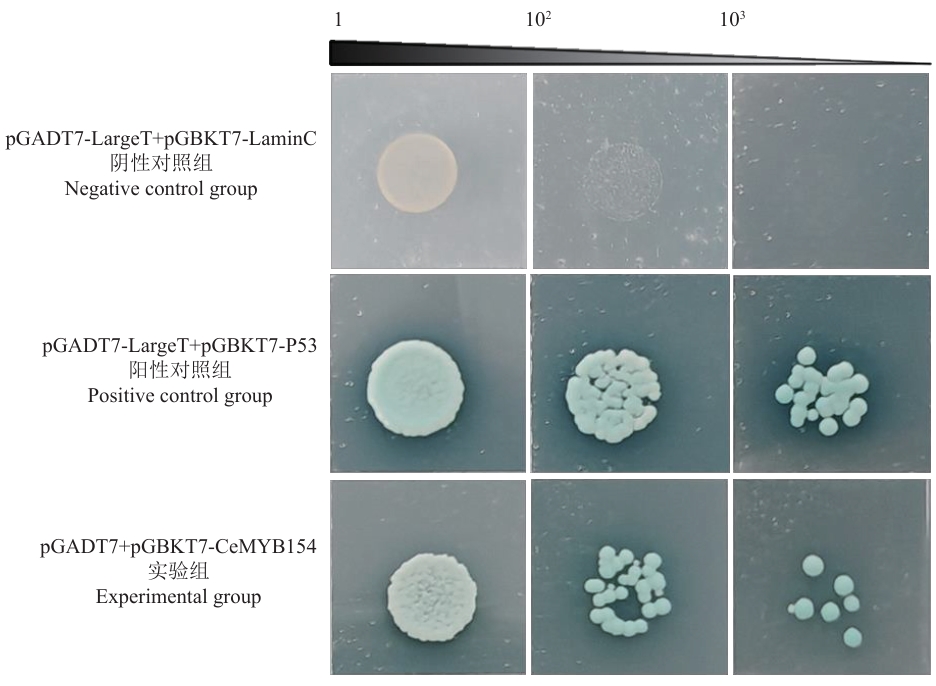
图3 CeMYB154转录激活活性鉴定1、102、103分别代表酵母菌液稀释倍数
Fig. 3 Identification of CeMYB154 transcriptional activation activity1, 102, 103 indicate the dilution multiple of yeast liquid, respectively
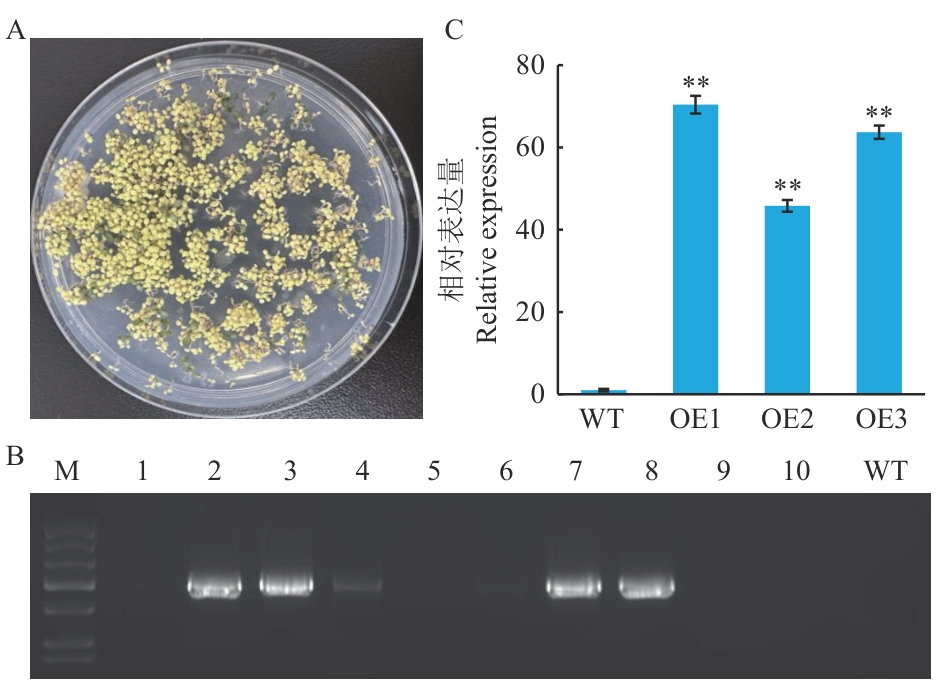
图4 转基因拟南芥植株的鉴定A:卡那霉素筛选T1转基因苗;B:T1转基因植株的PCR鉴定(M 为DL2000 marker,1‒10为转基因植株,WT为野生型植株);C:T2转基因拟南芥植株中CeMYB154的表达水平检测,OE1、OE2和OE3分别代表1个独立的T2代转基因植株,下同
Fig. 4 Identification of transgenic A. thaliana plantsA: Hygromycin screening T1 transgenic vaccines. B: PCR identification of T1 transgenic plants (M is DL2000 marker, 1‒10 are transgenic plants, and WT is wild-type plants). C: Detection of CeMYB154 expression in T2 transgenic A. thaliana plants, OE1, OE2 and OE3 indicate an independent T2 generation transgenic plant, respectively, the same below

图5 CeMYB154转基因拟南芥植株耐盐表型A:盐胁迫对拟南芥植株幼苗生长的影响;B:盐胁迫对转基因拟南芥植株根系生长的影响;C:盐胁迫下WT和转基因拟南芥植株的根长
Fig. 5 Phenotype of salt-tolerant in CeMYB154 transgenic A. thaliana plantsA: Effect of salt stress on the seedling growth of A. thaliana plants. B: Effect of salt stress on the root growth of transgenic A. thaliana plants. C: Root length of WT and transgenic A.thaliana plants under salt stress
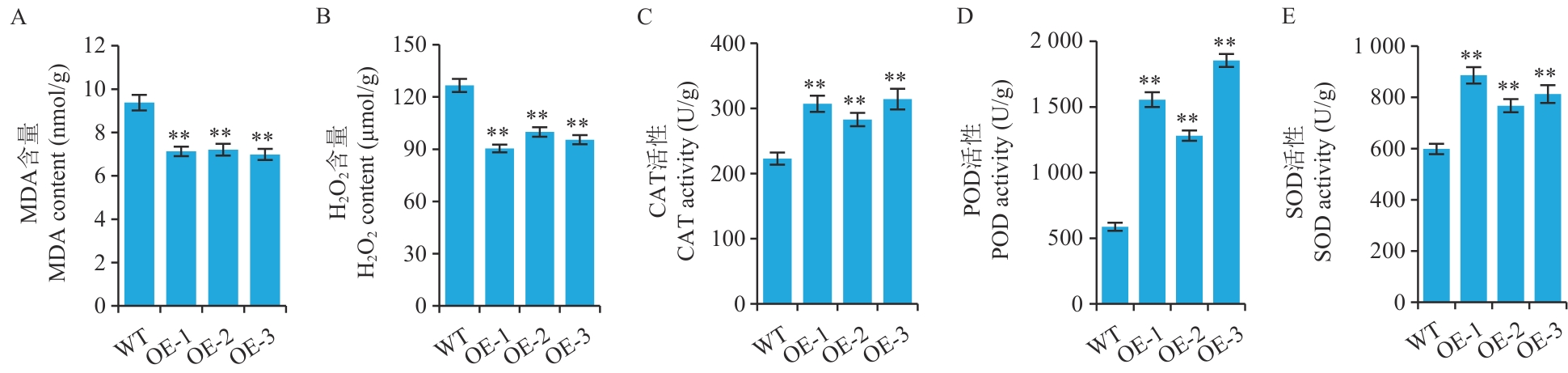
图6 盐胁迫下过表达CeMYB154拟南芥的相关生理指标测定
Fig. 6 Determination of physiological indexes related to the overexpression of CeMYB154 in A. thaliana under salt stress
| 1 | 张玉娟, 黎冬华, 宫慧慧, 等. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析 [J]. 生物技术通报, 2023, 39(11): 308-317. |
| Zhang YJ, Li DH, Gong HH, et al. Cloning and salt-tolerance analysis of NAC transcription factor SiNAC77 from Sesamum indicum L [J]. Biotechnol Bull, 2023, 39(11): 308-317. | |
| 2 | 杨帆, 朱文学. 油莎豆研究现状及展望 [J]. 粮食与油脂, 2020, 33(7): 4-6. |
| Yang F, Zhu WX. Research status and prospect of Cyperus esculentus [J]. Cereals Oils, 2020, 33(7): 4-6. | |
| 3 | 刘玉兰, 王小宁, 舒垚, 等. 不同产地油莎豆性状及组成分析研究 [J]. 中国油脂, 2020, 45(8): 125-129. |
| Liu YL, Wang XN, Shu Y, et al. Character and composition of Cyperus esculentus from different origins [J]. China Oils Fats, 2020, 45(8): 125-129. | |
| 4 | 王会伟, 张向歌, 李春鑫, 等. 油莎豆耐盐性评估及盐胁迫下幼苗根系转录组学分析 [J]. 作物学报, 2023, 49(7): 1882-1894. |
| Wang HW, Zhang XG, Li CX, et al. Evaluation of salt tolerance in Cyperus esculentus and transcriptomic analysis of seedling roots under salt stress [J]. Acta Agron Sin, 2023, 49(7): 1882-1894. | |
| 5 | 周扬, 王鹏, 李雨欣. 植物耐盐分子机制研究进展 [J]. 广东农业科学, 2023, 50(10): 97-109. |
| Zhou Y, Wang P, Li YX. Research progress on molecular mechanism of plant salt tolerance [J]. Guangdong Agric Sci, 2023, 50(10): 97-109. | |
| 6 | 冯凯月, 赵鑫焱, 李子妍, 等. 植物响应盐碱胁迫的分子机制研究进展 [J]. 生物技术通报, 2024, 40(10): 122-138. |
| Feng KY, Zhao XY, Li ZY, et al. Research advances in the molecular mechanisms of plant response to saline-alkali stress [J]. Biotechnol Bull, 2024, 40(10): 122-138. | |
| 23 | 李星伟, 王庆江, 吴宇童, 等. 苹果MdMYB113基因响应高盐胁迫的功能鉴定 [J]. 植物生理学报, 2021, 57(3): 579-588. |
| Li XW, Wang QJ, Wu YT, et al. Functional identification of MdMYB113 gene in apple in response to high salt stress [J]. Plant Physiol J, 2021, 57(3): 579-588. | |
| 24 | Cao YP, Han YH, Li DH, et al. MYB transcription factors in Chinese pear (Pyrus bretschneideri rehd.): genome-wide identification, classification, and expression profiling during fruit development [J]. Front Plant Sci, 2016, 7: 577. |
| 25 | Zhang ZJ, Zhang L, Liu Y, et al. Identification and expression analysis of R2R3-MYB family genes associated with salt tolerance in Cyclocarya paliurus [J]. Int J Mol Sci, 2022, 23(7): 3429. |
| 26 | 张红梅, 刘晓庆, 陈华涛, 等. 大豆转录因子GmWRKY58亚细胞定位及在非生物胁迫下的表达分析 [J]. 华北农学报, 2018, 33(2): 49-57. |
| Zhang HM, Liu XQ, Chen HT, et al. Subcellular localization and expression analysis of a soybean transcription factor GmWRKY58 in response to abiotic stresses [J]. Acta Agric Boreali Sin, 2018, 33(2): 49-57. | |
| 27 | 陈彧, 邢文婷, 李雨欣, 等. 铁皮石斛DcNAC1基因克隆、表达及转录自激活活性分析 [J]. 南方农业学报, 2023, 54(6): 1612-1621. |
| Chen Y, Xing WT, Li YX, et al. Cloning, expression and trans-activation activity analysis of DcNAC1 gene from Dendrobium catenatum [J]. J South Agric, 2023, 54(6): 1612-1621. | |
| 28 | Dossa K, Mmadi MA, Zhou R, et al. Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis [J]. AoB Plants, 2019, 12(1): plz081. |
| 29 | 胡雅丹, 伍国强, 刘晨, 等. MYB转录因子在调控植物响应逆境胁迫中的作用 [J]. 生物技术通报, 2024, 40(6): 5-22. |
| Hu YD, Wu GQ, Liu C, et al. Roles of MYB transcription factor in regulating the responses of plants to stress [J]. Biotechnol Bull, 2024, 40(6): 5-22. | |
| 30 | He YX, Dong YS, Yang XD, et al. Functional activation of a novel R2R3-MYB protein gene, GmMYB68, confers salt-alkali resistance in soybean (Glycine max L.) [J]. Genome, 2020, 63(1): 13-26. |
| 7 | Zhang S, Wu QR, Liu LL, et al. Osmotic stress alters circadian cytosolic Ca2+ oscillations and OSCA1 is required in circadian gated stress adaptation [J]. Plant Signal Behav, 2020, 15(12): 1836883. |
| 8 | Jiang ZH, Zhou XP, Tao M, et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx [J]. Nature, 2019, 572(7769): 341-346. |
| 9 | Chen K, Gao JH, Sun SJ, et al. BONZAI proteins control global osmotic stress responses in plants [J]. Curr Biol, 2020, 30(24): 4815-4825.e4. |
| 10 | Yan JW, Liu Y, Yan JW, et al. The salt-activated CBF1/CBF2/CBF3-GALS1 module fine-tunes galactan-induced salt hypersensitivity in Arabidopsis [J]. J Integr Plant Biol, 2023, 65(8): 1904-1917. |
| 11 | Gao F, Zhou J, Deng RY, et al. Overexpression of a Tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis [J]. J Plant Physiol, 2017, 214: 81-90. |
| 12 | Le Hir R, Castelain M, Chakraborti D, et al. AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana [J]. Physiol Plant, 2017, 160(3): 312-327. |
| 13 | Raineri J, Wang SH, Peleg Z, et al. The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress [J]. Plant Mol Biol, 2015, 88(4/5): 401-413. |
| 14 | Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis [J]. Trends Plant Sci, 2010, 15(10): 573-581. |
| 15 | Wang YJ, Zhang Y, Fan CJ, et al. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia [J]. BMC Plant Biol, 2021, 21(1): 328. |
| 16 | Liao Y, Zou HF, Wang HW, et al. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants [J]. Cell Res, 2008, 18(10): 1047-1060. |
| 17 | Yoo JH, Park CY, Kim JC, et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis [J]. J Biol Chem, 2005, 280(5): 3697-3706. |
| 18 | Zhu N, Cheng SF, Liu XY, et al. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice [J]. Plant Sci, 2015, 236: 146-156. |
| 19 | Zhang X, Chen LC, Shi QH, et al. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato [J]. Plant Sci, 2020, 291: 110356. |
| 20 | Zhao YY, Yang ZE, Ding YP, et al. Over-expression of an R2R3 MYB gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis [J]. Plant Sci, 2019, 286: 28-36. |
| 21 | Dong W, Liu XJ, Li DL, et al. Transcriptional profiling reveals that a MYB transcription factor MsMYB4 contributes to the salinity stress response of alfalfa [J]. PLoS One, 2018, 13(9): e0204033. |
| 22 | Du BY, Liu H, Dong KT, et al. Over-expression of an R2R3 MYB gene, MdMYB108L, enhances tolerance to salt stress in transgenic plants [J]. Int J Mol Sci, 2022, 23(16): 9428. |
| 31 | Zhang WX, Wang N, Yang JT, et al. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean [J]. Plant Sci, 2020, 291: 110326. |
| 32 | Du YT, Zhao MJ, Wang CT, et al. Identification and characterization of GmMYB118 responses to drought and salt stress [J]. BMC Plant Biol, 2018, 18(1): 320. |
| 33 | Pi EX, Xu J, Li HH, et al. Enhanced salt tolerance of rhizobia-inoculated soybean correlates with decreased phosphorylation of the transcription factor GmMYB183 and altered flavonoid biosynthesis [J]. Mol Cell Proteomics, 2019, 18(11): 2225-2243. |
| 34 | Pi EX, Zhu CM, Fan W, et al. Quantitative phosphoproteomic and metabolomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt stress [J]. Mol Cell Proteomics, 2018, 17(6): 1209-1224. |
| 35 | Wang C, Wang LJ, Lei J, et al. IbMYB308, a sweet potato R2R3-MYB gene, improves salt stress tolerance in transgenic tobacco [J]. Genes, 2022, 13(8): 1476. |
| 36 | Hossain MA, Bhattacharjee S, Armin SM, et al. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging [J]. Front Plant Sci, 2015, 6: 420. |
| 37 | Liu P, Wu XL, Gong BB, et al. Review of the mechanisms by which transcription factors and exogenous substances regulate ROS metabolism under abiotic stress [J]. Antioxidants, 2022, 11(11): 2106. |
| 38 | Li WH, Zhong JL, Zhang LH, et al. Overexpression of a Fragaria vesca MYB transcription factor gene (FvMYB82) increases salt and cold tolerance in Arabidopsis thaliana [J]. Int J Mol Sci, 2022, 23(18): 10538. |
| 39 | 程钰鑫. R2R3 MYB转录因子MYB71、MYB79和MYB121对拟南芥ABA及干旱胁迫响应的调控 [D]. 长春: 东北师范大学, 2022. |
| Cheng YX. Regulation of R2R3 MYB transcription factors MYB71, MYB79 and MYB121 on ABA and drought stress response in Arabidopsis thaliana [D]. Changchun: Northeast Normal University, 2022. |
| [1] | 许慧珍, SHANTWANA Ghimire, RAJU Kharel, 岳云, 司怀军, 唐勋. 马铃薯SUMO E3连接酶基因家族分析及StSIZ1基因的克隆与表达模式分析[J]. 生物技术通报, 2025, 41(6): 119-129. |
| [2] | 吴娅, 姚润, 杨含婷, 刘微, 杨帅, 宋驰, 陈士林. 凤梨薄荷SDR基因家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(5): 175-185. |
| [3] | 李志强, 罗正乾, 徐琳黎, 周国慧, 屈丝雨, 刘恩良, 顼东婷. 基于T2T基因组鉴定大豆R2R3-MYB基因家族及干旱和盐胁迫下的表达分析[J]. 生物技术通报, 2025, 41(5): 141-152. |
| [4] | 杨春, 王晓倩, 王红军, 晁跃辉. 蒺藜苜蓿MtZHD4基因克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2025, 41(5): 244-254. |
| [5] | 宋慧洋, 苏宝杰, 李京昊, 梅超, 宋倩娜, 崔福柱, 冯瑞云. 马铃薯StAS2-15基因的克隆及盐胁迫下功能分析[J]. 生物技术通报, 2025, 41(5): 119-128. |
| [6] | 班秋艳, 赵鑫月, 迟文静, 黎俊生, 王琼, 夏瑶, 梁丽云, 贺巍, 李叶云, 赵广山. 茶树光敏色素互作因子CsPIF3a的克隆及其与光温逆境的响应[J]. 生物技术通报, 2025, 41(4): 256-265. |
| [7] | 卢勇杰, 夏海乾, 李永铃, 张文建, 余婧, 赵会纳, 王兵, 许本波, 雷波. 烟草AP2/ERF转录因子NtESR2的克隆及功能分析[J]. 生物技术通报, 2025, 41(4): 266-277. |
| [8] | 佟德利, 张馨, 陈佳庆, 贺海升. 蓝莓内生细菌的分离及其对上海青铝胁迫的缓解作用[J]. 生物技术通报, 2025, 41(4): 302-311. |
| [9] | 刘涛, 王志淇, 吴文博, 石文婷, 王超楠, 杜崇, 杨中敏. 马铃薯GRAM基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(4): 145-155. |
| [10] | 钱祺, 王增辉, 孙荣华, 罗英智, 苏良辰. 花生蛋白磷酸酶AhPDCP37的抗旱性功能研究[J]. 生物技术通报, 2025, 41(3): 98-103. |
| [11] | 李彩霞, 李艺, 穆宏秀, 林俊轩, 白龙强, 孙美华, 苗妍秀. 中国南瓜bHLH转录因子家族的鉴定与生物信息学分析[J]. 生物技术通报, 2025, 41(1): 186-197. |
| [12] | 袁柳娇, 黄文琳, 陈崇志, 梁敏, 黄梓淇, 陈雪雪, 陈日檬, 王锂韫. 盐胁迫对广藿香叶片生理特性、超微结构及药效成分的影响[J]. 生物技术通报, 2025, 41(1): 230-239. |
| [13] | 乔岩, 杨芳, 任盼荣, 祁伟亮, 安沛沛, 李茜, 李丹, 肖俊飞. 马铃薯野生种烯酰水合酶超家族基因ScDHNS的克隆与功能分析[J]. 生物技术通报, 2024, 40(9): 92-103. |
| [14] | 邢丽南, 张艳芳, 葛明然, 赵令敏, 陈妍, 霍秀文. 山药DoWRKY40基因表达特征分析及互作蛋白筛选[J]. 生物技术通报, 2024, 40(8): 118-128. |
| [15] | 武帅, 辛燕妮, 买春海, 穆晓娅, 王敏, 岳爱琴, 赵晋忠, 吴慎杰, 杜维俊, 王利祥. 大豆GS基因家族全基因组鉴定及胁迫响应分析[J]. 生物技术通报, 2024, 40(8): 63-73. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||