








生物技术通报 ›› 2025, Vol. 41 ›› Issue (9): 242-255.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0202
• 研究报告 • 上一篇
程婷婷1( ), 刘俊1(
), 刘俊1( ), 王利丽1, 练从龙1, 魏文君1, 郭辉1, 吴尧琳2, 杨晶凡1, 兰金旭1, 陈随清1,3
), 王利丽1, 练从龙1, 魏文君1, 郭辉1, 吴尧琳2, 杨晶凡1, 兰金旭1, 陈随清1,3
收稿日期:2025-02-25
出版日期:2025-09-26
发布日期:2025-09-24
通讯作者:
刘俊,女,博士,助理研究员,研究方向 :药用植物生理生态及分子生物学;E-mail: liujun_0325@163.com作者简介:程婷婷,女,硕士研究生,研究方向 :药用植物分子生物学;E-mail: 19503888810@163.com
基金资助:
CHENG Ting-ting1( ), LIU Jun1(
), LIU Jun1( ), WANG Li-li1, LIAN Cong-long1, WEI Wen-jun1, GUO Hui1, WU Yao-lin2, YANG Jing-fan1, LAN Jin-xu1, CHEN Sui-qing1,3
), WANG Li-li1, LIAN Cong-long1, WEI Wen-jun1, GUO Hui1, WU Yao-lin2, YANG Jing-fan1, LAN Jin-xu1, CHEN Sui-qing1,3
Received:2025-02-25
Published:2025-09-26
Online:2025-09-24
摘要:
目的 查尔酮异构酶(CHI)是类黄酮生物合成途径中的关键限速酶。对杜仲CHI基因家族成员进行全基因组鉴定,系统分析其表达特征,以探究其在杜仲生长发育及逆境响应中的生物学功能。 方法 基于杜仲基因组数据,采用生物信息学方法系统鉴定CHI基因家族成员,并全面分析其理化性质、染色体定位、启动子顺式作用元件、进化关系、保守基序和基因结构等特征。同时,结合转录组数据和RT-qPCR技术,探究EuCHIs在不同组织及非生物胁迫处理下的表达模式。 结果 在杜仲基因组中共鉴定出7个EuCHI基因家族成员(EuCHI-1-EuCHI-7),其编码蛋白的氨基酸长度为84-388 aa,分子量为9 473.24-43 248.30 Da,等电点范围为4.81-9.42,不稳定指数范围为23.09-46.43,亲疏水性范围为-0.381-0.206,不均匀地分布在5条染色体上。亚细胞定位预测显示,除EuCHI-2定位于细胞核中;其余成员均定位于叶绿体中,此外,EuCHI-1在细胞膜中也有分布,EuCHI-4在细胞质和线粒体中也有分布。系统进化分析将EuCHIs划分为Group Ⅰ、Group Ⅱ和Group Ⅵ 3个亚家族,各成员含有1-10个外显子。启动子分析表明,EuCHIs可能参与生长发育和激素响应等生物学过程,转录组数据表明大部分EuCHIs基因参与杜仲叶色形成以及花发育等多种生物学过程。RT-qPCR结果显示,EuCHIs具有组织表达特异性,除EuCHI-3外,其余均在根部高量表达;与ABA和GA3胁迫处理相比,干旱胁迫处理更能显著诱导EuCHI基因的表达。 结论 鉴定出7个EuCHIs基因,分为3个亚家族,EuCHIs在杜仲不同组织和不同胁迫中表达模式存在差异,表明它们在杜仲生长发育和非生物胁迫中发挥重要作用。
程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255.
CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns[J]. Biotechnology Bulletin, 2025, 41(9): 242-255.
基因名称 Gene name | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| EuCHI-1 | TCAGAAGTGCAAAACGATGC | GTCGGGCCAAGTCAAAAATA |
| EuCHI-2 | TGCTGATCACGAGAAACTGG | CGGTGAGTTTCCTGATGGAT |
| EuCHI-3 | ACTGGAACCTACACCGATGC | GCCCAATTTCAGGAAATGAA |
| EuCHI-4 | AATTCGATGTCCAAGCGTTC | ACGGACTCCATCAACTCCTC |
| EuCHI-5 | CCGTCGTCACTACCTCCAAT | GAAACGGGACAGCAACAAAT |
| EuCHI-6 | TTTCCTCCGGAGATCAACAC | TTTTCAACTGGGGCAGAAAC |
| EuCHI-7 | GAGCCTATTACGGGCATTGA | TCATGGCATTTGTTCAGCTC |
| UBC | AGTGGGTGGTGCTGTAGTCC | AACTCCCGTTTCGTTTGTTG |
表1 EuCHIs基因引物序列
Table 1 Primer sequences of EuCHIs genes
基因名称 Gene name | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| EuCHI-1 | TCAGAAGTGCAAAACGATGC | GTCGGGCCAAGTCAAAAATA |
| EuCHI-2 | TGCTGATCACGAGAAACTGG | CGGTGAGTTTCCTGATGGAT |
| EuCHI-3 | ACTGGAACCTACACCGATGC | GCCCAATTTCAGGAAATGAA |
| EuCHI-4 | AATTCGATGTCCAAGCGTTC | ACGGACTCCATCAACTCCTC |
| EuCHI-5 | CCGTCGTCACTACCTCCAAT | GAAACGGGACAGCAACAAAT |
| EuCHI-6 | TTTCCTCCGGAGATCAACAC | TTTTCAACTGGGGCAGAAAC |
| EuCHI-7 | GAGCCTATTACGGGCATTGA | TCATGGCATTTGTTCAGCTC |
| UBC | AGTGGGTGGTGCTGTAGTCC | AACTCCCGTTTCGTTTGTTG |
基因ID Gene ID | 基因名 Gene name | 氨基酸数量Number of amino acids (aa) | 分子量 Molecular weight (Da) | 等电点 Isoelectric point pI | 不稳定指数 Instability index | 亚细胞定位 Subcellular localization | 亲水性总平均值 Hydrophilicity and hydrophobicity |
|---|---|---|---|---|---|---|---|
| EUC25438-RA | EuCHI-1 | 291 | 31 099.58 | 9.28 | 36.93 | 细胞膜、叶绿体 | 0.047 |
| EUC24475-RA | EuCHI-2 | 280 | 30 689.55 | 6.29 | 35.13 | 细胞核 | -0.381 |
| EUC08195-RA | EuCHI-3 | 259 | 27 648.28 | 4.81 | 30.60 | 叶绿体 | -0.086 |
| EUC08196-RA | EuCHI-4 | 84 | 9 473.24 | 9.42 | 23.09 | 叶绿体、细胞质、线粒体 | 0.206 |
| EUC26357-RA | EuCHI-5 | 283 | 30 748.30 | 9.17 | 46.43 | 叶绿体 | -0.077 |
| EUC24815-RA | EuCHI-6 | 203 | 22 629.79 | 4.85 | 33.83 | 叶绿体 | -0.144 |
| EUC14152-RA | EuCHI-7 | 388 | 43 248.30 | 8.22 | 44.54 | 叶绿体 | -0.146 |
表2 杜仲CHIs蛋白的理化性质分析
Table 2 Analysis of physical and chemical properties of CHI proteins from E. ulmoides
基因ID Gene ID | 基因名 Gene name | 氨基酸数量Number of amino acids (aa) | 分子量 Molecular weight (Da) | 等电点 Isoelectric point pI | 不稳定指数 Instability index | 亚细胞定位 Subcellular localization | 亲水性总平均值 Hydrophilicity and hydrophobicity |
|---|---|---|---|---|---|---|---|
| EUC25438-RA | EuCHI-1 | 291 | 31 099.58 | 9.28 | 36.93 | 细胞膜、叶绿体 | 0.047 |
| EUC24475-RA | EuCHI-2 | 280 | 30 689.55 | 6.29 | 35.13 | 细胞核 | -0.381 |
| EUC08195-RA | EuCHI-3 | 259 | 27 648.28 | 4.81 | 30.60 | 叶绿体 | -0.086 |
| EUC08196-RA | EuCHI-4 | 84 | 9 473.24 | 9.42 | 23.09 | 叶绿体、细胞质、线粒体 | 0.206 |
| EUC26357-RA | EuCHI-5 | 283 | 30 748.30 | 9.17 | 46.43 | 叶绿体 | -0.077 |
| EUC24815-RA | EuCHI-6 | 203 | 22 629.79 | 4.85 | 33.83 | 叶绿体 | -0.144 |
| EUC14152-RA | EuCHI-7 | 388 | 43 248.30 | 8.22 | 44.54 | 叶绿体 | -0.146 |
基因名 Gene name | α-螺旋α-helix | 延伸链Extended chain | 无规则卷曲Random coil | |||
|---|---|---|---|---|---|---|
数量 Quantity | 占比 Proportion (%) | 数量 Quantity | 占比 Proportion (%) | 数量 Quantity | 占比 Proportion (%) | |
| EuCHI-1 | 96 | 32.99 | 48 | 16.49 | 147 | 50.52 |
| EuCHI-2 | 109 | 38.93 | 41 | 14.64 | 130 | 46.43 |
| EuCHI-3 | 108 | 41.70 | 45 | 17.37 | 106 | 40.93 |
| EuCHI-4 | 31 | 36.90 | 22 | 26.19 | 31 | 36.90 |
| EuCHI-5 | 101 | 35.69 | 44 | 15.55 | 138 | 48.76 |
| EuCHI-6 | 87 | 42.86 | 41 | 20.20 | 75 | 36.95 |
| EuCHI-7 | 126 | 32.47 | 58 | 14.95 | 204 | 52.58 |
表3 杜仲CHIs基因家族二级结构分析
Table 3 Secondary structure analysis of E. ulmoidesCHIs gene family
基因名 Gene name | α-螺旋α-helix | 延伸链Extended chain | 无规则卷曲Random coil | |||
|---|---|---|---|---|---|---|
数量 Quantity | 占比 Proportion (%) | 数量 Quantity | 占比 Proportion (%) | 数量 Quantity | 占比 Proportion (%) | |
| EuCHI-1 | 96 | 32.99 | 48 | 16.49 | 147 | 50.52 |
| EuCHI-2 | 109 | 38.93 | 41 | 14.64 | 130 | 46.43 |
| EuCHI-3 | 108 | 41.70 | 45 | 17.37 | 106 | 40.93 |
| EuCHI-4 | 31 | 36.90 | 22 | 26.19 | 31 | 36.90 |
| EuCHI-5 | 101 | 35.69 | 44 | 15.55 | 138 | 48.76 |
| EuCHI-6 | 87 | 42.86 | 41 | 20.20 | 75 | 36.95 |
| EuCHI-7 | 126 | 32.47 | 58 | 14.95 | 204 | 52.58 |

图2 杜仲CHIs基因启动子元件分析A:EuCHIs基因启动子元件定位图;B:EuCHIs基因中每个元件数量图;C:EuCHIs基因每类元件总数图;D:3类元件(从左至右依次为激素响应元件、胁迫响应元件和生长发育响应元件)中各个元件占比图
Fig. 2 Analysis of promoter elements of E. ulmoides CHIs genesA: Location map of promoter elements of EuCHIs genes. B: Number map of each elements in EuCHIs genes. C: Total number map of each type of elements in EuCHIs genes. D: Proportion map of each element in three categories of elements (From left to right are hormone response element, stress response element, growth and development response element)

图5 EuCHIs保守基序和基因结构分析A:EuCHIs保守基序;B:EuCHIs基因结构;C:保守基序氨基酸序列
Fig. 5 Analysis of conserved motifs and gene structures of EuCHIsA: Conserved motifs of EuCHIs. B: Gene structure of EuCHIs. C: Conserved motif amino acid sequence
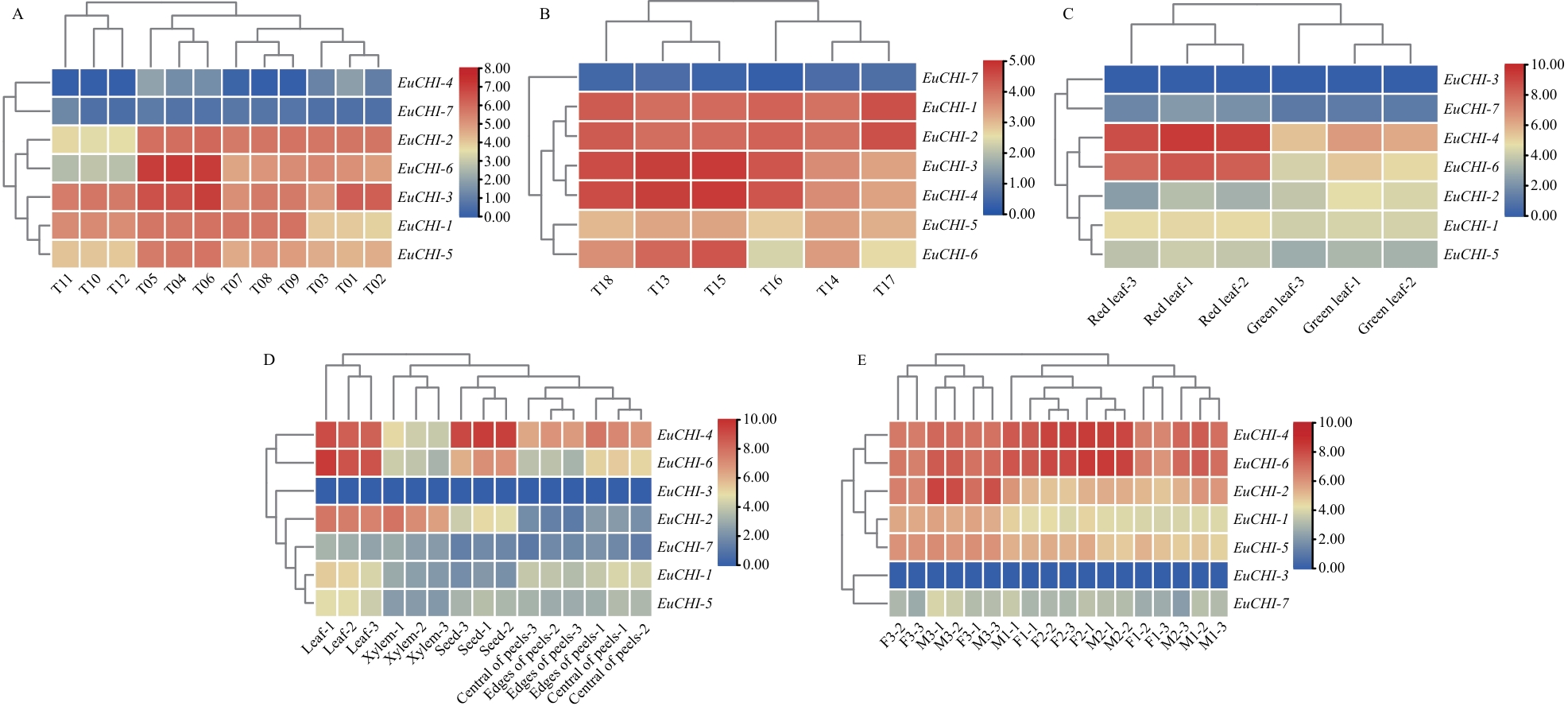
图7 杜仲CHIs基因表达模式A:叶片不同发育时期表达模式分析(T01-T03叶芽;T04-T06初生叶;T07-T09幼叶;T10-T12老叶);B:低和高胶含量叶片的表达模式(T13-T15低胶含量叶片;T16-T18高胶含量叶片);C:红叶和绿叶的表达模式;D:不同组织的表达模式分析;E:不同花发育的表达模式分析(F1:雌花形态分化初期;F2:雌花诱导期;F3:雌花成熟期;M1:雄花形态分化初期;M2:雄花诱导期;M3:雄花成熟期)
Fig. 7 Expression patterns of E. ulmoidesCHIs genesA: Analysis of expression patterns at different leaf development stages (T01-T03: Leaf buds. T04-T06: Primary leaves. T07-T09: Young leaves. T10-T12: Old leaves). B: Expression patterns of leaves with low and high gum contents (T13-T15: Leaves with low gum content. T16-T18: Leaves with high gum content). C: Expression patterns of red and green leaves. D: Analysis of expression patterns in different tissues. E: Analysis of expression patterns during different flower development stages (F1: Early stage of female flower morphological differentiation. F2: Induction stage of female flowers. F3: Maturation stage of female flowers. M1: Early stage of male flower morphological differentiation. M2: Induction stage of male flowers. M3: Maturation stage of male flowers)
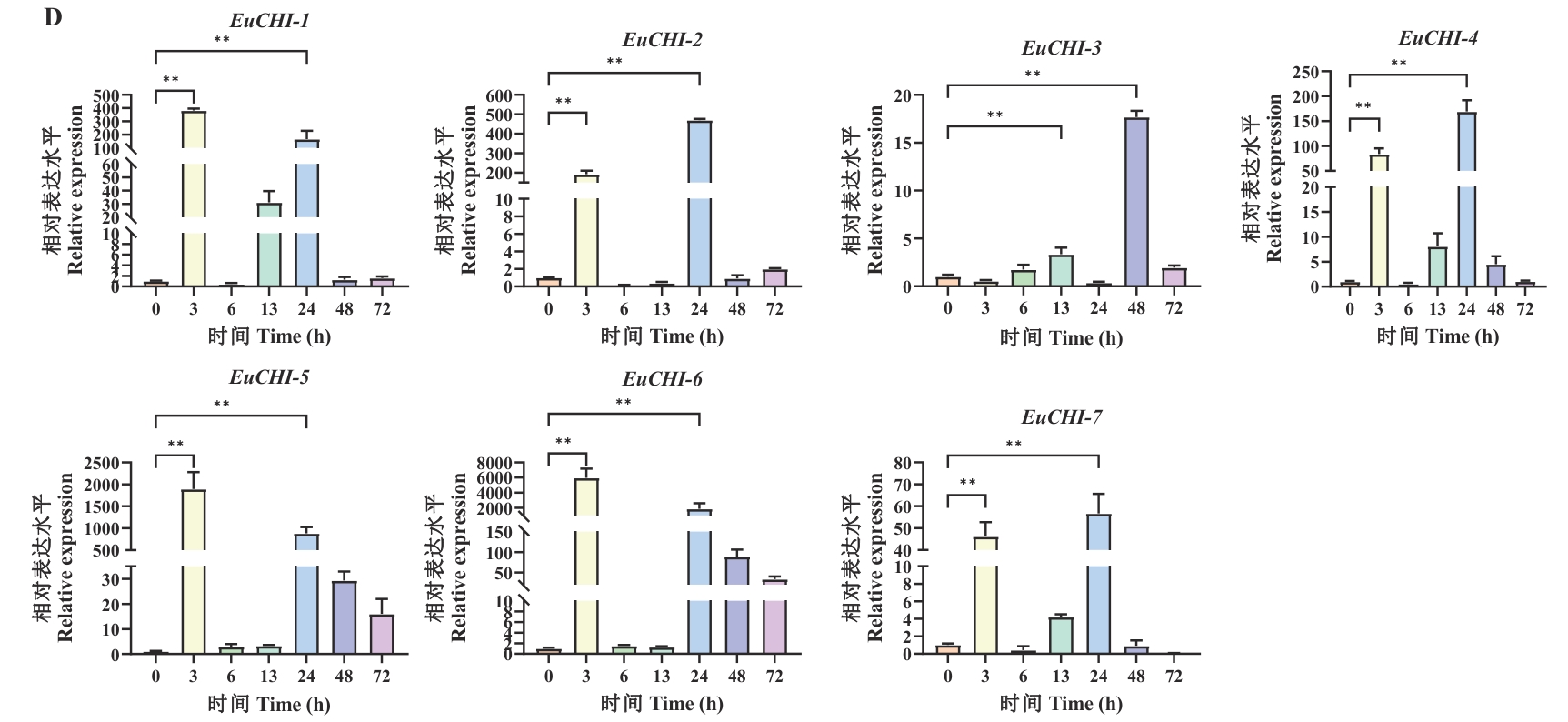
图8 EuCHIs表达基因的RT-qPCR分析A:EuCHIs基因在不同组织中的表达模式;与根相比,*P<0.05,**P<0.01;与茎相比,##P<0.01;B:EuCHIs基因在ABA处理中的表达模式;与0 h相比,*P<0.05,**P<0.01;C:EuCHIs基因在GA3处理中的表达模式;与0 h相比,*P<0.05,**P<0.01;D:EuCHIs基因在干旱处理中的表达模式;与0 h相比,**P<0.01
Fig. 8 RT-qPCR analysis of the EuCHIs expressed genesA: Expression patterns of the EuCHIs genes in different tissues. * P<0.05 and ** P<0.01 compared with roots; ##P<0.01 compared with stems. B: Expression patterns of the EuCHIs genes during ABA treatment. *P<0.05 and **P<0.01 compared with 0 h. C: Expression patterns of the EuCHIs genes during GA3 treatment. *P<0.05 and **P<0.01 compared with 0 h. D: Expression patterns of the EuCHIs genes under drought treatment. **P<0.01 compared with 0 h
| [1] | 高宏伟, 李玉萍, 李守超. 杜仲的化学成分及药理作用研究进展 [J]. 中医药信息, 2021, 38(6): 73-81. |
| Gao HW, Li YP, Li SC. Study advances in chemical constituents and pharmacological effects of Eucommia ulmoides Oliv [J]. Inf Tradit Chin Med, 2021, 38(6): 73-81. | |
| [2] | 贾征, 黄文, 薛安. 杜仲叶黄酮的超声提取及其抗氧化性研究 [J]. 安徽农业科学, 2008, 36(4): 1286-1288. |
| Jia Z, Huang W, Xue A. Extraction of flavonoids from Eucommia ulmoides leaves by ultrasonic and research on its antioxidation [J]. J Anhui Agric Sci, 2008, 36(4): 1286-1288. | |
| [3] | 李天来, 卢银让, 潘建平, 等. 杜仲叶茶功效成分对小鼠免疫功能影响 [J]. 中国公共卫生, 2007, 23(10): 1221-1223. |
| Li TL, Lu YR, Pan JP, et al. Effects of Eucommia ulmoides Oliv.leaves tea on mouse immunity function [J]. Chin J Public Health, 2007, 23(10): 1221-1223. | |
| [4] | 王梦华, 刘颖菊, 周歧新, 等. 杜仲叶乙醇提取物抗动脉粥样硬化作用的实验研究 [J]. 中成药, 2007, 29(11): 1687-1690. |
| Wang MH, Liu YJ, Zhou QX, et al. Experimental study on anti-atherosclerosis effect of ethanol extract from Eucommia ulmoides leaves [J]. Chin Tradit Pat Med, 2007, 29(11): 1687-1690. | |
| [5] | 孙兰萍, 马龙, 张斌, 等. 杜仲黄酮类化合物的研究进展 [J]. 食品工业科技, 2009, 30(3): 359-363. |
| Sun LP, Ma L, Zhang B, et al. Research progress of flavonoids in Eucommia ulmoides [J]. Sci Technol Food Ind, 2009, 30(3): 359-363. | |
| [6] | 叶力, 谢笔钧, 胡慰望. 杜仲叶中黄酮类化合物的研究 [J]. 中草药, 1998, 29(11): 746-747. |
| Ye L, Xie BJ, Hu WW. Study on flavonoid compounds in Eucommia ulmoides Oliv. leaves [J]. Chin Tradit Herb Drugs, 1998, 29(11): 746-747. | |
| [7] | Kim HY, Moon BH, Lee HJ, et al. Flavonol glycosides from the leaves of Eucommia ulmoides with glycation inhibitory activity [J]. J Ethnopharmacol, 2004, 93(2/3): 227-230. |
| [8] | 赵德义, 高锦明, 许爱遐, 等. 杜仲黄酮指纹图谱研究 [J]. 西北植物学报, 2003, 23(11): 1987-1989. |
| Zhao DY, Gao JM, Xu AX, et al. Study on fingerprint analysis and bioactive functions of Eucommia ulmoides flavonoids [J]. Acta Bot Boreali Occidentalia Sin, 2003, 23(11): 1987-1989. | |
| [9] | Sun YX, Zhang AD, Landis JB, et al. Genome assembly of the snow Lotus species Saussurea involucrata provides insights into acacetin and rutin biosynthesis and tolerance to an alpine environment [J]. Hortic Res, 2023, 10(10): uhad180. |
| [10] | Sun W, Shen H, Xu H, et al. Chalcone isomerase a key enzyme for anthocyanin biosynthesis in Ophiorrhiza japonica [J]. Front Plant Sci, 2019, 10: 865. |
| [11] | 张杨青慧. 菊花衰老过程花瓣变色机理初步研究 [D]. 南京: 南京农业大学, 2017. |
| Zhang Y. Preliminary study on mechanism of floral coloration during senescent process of chrysnathemum moricolium [D]. Nanjing: Nanjing Agricultural University, 2017. | |
| [12] | 王仁杰, 刘雄盛, 李梦婷, 等. 基于代谢组学的枫香叶片变红机理研究 [J]. 广西林业科学, 2024, 53(4): 514-522. |
| Wang RJ, Liu XS, Li MT, et al. Study on reddening mechanism of Liquidambar formosana leaves based on metabolomics [J]. Guangxi For Sci, 2024, 53(4): 514-522. | |
| [13] | Li A, Wang FX, Ding TT, et al. Genome-wide DNA methylation dynamics and RNA-seq analysis during grape (cv. ‘Cabernet Franc’) skin coloration [J]. Genomics, 2024, 116(2): 110810. |
| [14] | 任超翔, 唐小慧, 何雯, 等. 红花查尔酮异构酶基因的克隆及表达分析 [J]. 天然产物研究与开发, 2018, 30(9): 1521-1525, 1574. |
| Ren CX, Tang XH, He W, et al. Cloning and expression analysis of Chalcone isomerase gene in safflower [J]. Nat Prod Res Dev, 2018, 30(9): 1521-1525, 1574. | |
| [15] | 薛永常, 麻新妍. 黄芩查尔酮异构酶基因的全长cDNA克隆及生物信息学分析 [J]. 中国野生植物资源, 2022, 41(6): 50-53, 96. |
| Xue YC, Ma XY. Cloning and bioinformatic analysis of Chalcone isomerase gene full-length cDNA from Scutellaria baicalensis [J]. Chin Wild Plant Resour, 2022, 41(6): 50-53, 96. | |
| [16] | Nishihara M, Nakatsuka T, Yamamura S. Flavonoid components and flower color change in transgenic tobacco plants by suppression of Chalcone isomerase gene [J]. FEBS Lett, 2005, 579(27): 6074-6078. |
| [17] | Mehdy MC, Lamb CJ. Chalcone isomerase cDNA cloning and mRNA induction by fungal elicitor, wounding and infection [J]. EMBO J, 1987, 6(6): 1527-1533. |
| [18] | van Tunen AJ, Koes RE, Spelt CE, et al. Cloning of the two Chalcone flavanone isomerase genes from Petunia hybrida: coordinate, light-regulated and differential expression of flavonoid genes [J]. EMBO J, 1988, 7(5): 1257-1263. |
| [19] | 荐红举, 万梦圆, 付毅, 等. 马铃薯CHI家族基因鉴定及其对外源水杨酸和茉莉酸的响应分析 [J]. 园艺学报, 2021, 48(1): 83-95. |
| Jian HJ, Wan MY, Fu Y, et al. Genome-wide identification and expression analysis of chitinase gene family in potato and its response to exogenous salicylic acid and jasmonic acid [J]. Acta Hortic Sin, 2021, 48(1): 83-95. | |
| [20] | Yu SH, Li JY, Peng T, et al. Identification of chalcone isomerase family genes and roles of CnCHI4 in flavonoid metabolism in Camellia nitidissima [J]. Biomolecules, 2022, 13(1): 41. |
| [21] | 吴倩倩, 夏玲, 于飞, 等. 巴西橡胶树查尔酮异构酶HbCHI基因家族的鉴定和基因功能初步验证 [J]. 热带作物学报, 2025, 46(2): 247-255. |
| Wu QQ, Xia L, Yu F, et al. Identification and preliminary verification of gene function of Chalcone isomerase HbCHI gene family in Hevea brasiliensis [J]. Chin J Trop Crops, 2025, 46(2): 247-255. | |
| [22] | 杨丹, 鲍军秀, 陈沛, 等. 棉花查尔酮异构酶基因家族的鉴定及表达分析 [J]. 华中农业大学学报, 2023, 42(5): 94-104. |
| Yang D, Bao JX, Chen P, et al. Identification and expression analysis of chalcone isomerase gene family in cotton [J]. J Huazhong Agric Univ, 2023, 42(5): 94-104. | |
| [23] | 翟妞, 郑庆霞, 刘萍萍, 等. 烟草查尔酮异构酶基因2(NtCHI2)功能分析 [J]. 烟草科技, 2020, 53(9): 1-5. |
| Zhai N, Zheng QX, Liu PP, et al. Functional analysis of chalcone isomerase gene 2 (NtCHI2) in tobacco [J]. Tob Sci Technol, 2020, 53(9): 1-5. | |
| [24] | 刘俊, 李龙, 陈玉龙, 等. 杜仲LBD基因家族全基因组鉴定与进化表达分析 [J]. 中草药, 2022, 53(10): 3142-3155. |
| Liu J, Li L, Chen YL, et al. Identification, evolution and expression analysis of LBD gene family in Eucommia ulmoides [J]. Chin Tradit Herb Drugs, 2022, 53(10): 3142-3155. | |
| [25] | 刘俊, 李龙, 陈玉龙, 等. 杜仲WOX家族基因鉴定及在叶片发育中的表达 [J]. 浙江农林大学学报, 2023, 40(1): 1-11. |
| Liu J, Li L, Chen YL, et al. Identification of WOX gene family and their expression in the leaf development of Eucommia ulmoides [J]. J Zhejiang A F Univ, 2023, 40(1): 1-11. | |
| [26] | Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0 [J]. Mol Biol Evol, 2013, 30(12): 2725-2729. |
| [27] | Kim S, Jones R, Yoo KS, et al. Gold color in Onions (Allium cepa): a natural mutation of the Chalcone isomerase gene resulting in a premature stop codon [J]. Mol Genet Genomics, 2004, 272(4): 411-419. |
| [28] | 路买林, 陈梦娇, 张嘉嘉, 等.'红叶'杜仲叶色转变过程中叶片生理指标变化 [J]. 南京林业大学学报: 自然科学版, 2021, 45(1): 86-92. |
| Lu ML, Chen MJ, Zhang JJ, et al. Leaf physiological indicator changes in the transformation of leaves color of Eucommia ulmoides ‘Hongye’ [J]. J Nanjing For Univ Nat Sci Ed, 2021, 45(1): 86-92. | |
| [29] | 徐朝阳, 张苛苛, 王晨晨, 等. 生菜查尔酮异构酶基因克隆及体外酶活测定 [J]. 植物生理学报, 2023, 59(11): 2135-2143. |
| Xu ZY, Zhang KK, Wang CC, et al. Cloning and catalytic analysis of Chalcone isomerase in vitro in lettuce (Lactuca sativa) [J]. Plant Physiol J, 2023, 59(11): 2135-2143. | |
| [30] | 雒晓鹏, 白悦辰, 高飞, 等. 金荞麦查尔酮异构酶的基因克隆及其花期表达与黄酮量分析 [J]. 中草药, 2013, 44(11): 1481-1485. |
| Luo XP, Bai YC, Gao F, et al. Gene cloning and expression level of chalcone isomerase during florescence and content of flavonoids in Fagopyrum dibotrys [J]. Chin Tradit Herb Drugs, 2013, 44(11): 1481-1485. | |
| [31] | Lambais MR. Suppression of endochitinase, β-1,3-endoglucanase, and Chalcone isomerase expression in bean vesicular-arbuscular mycorrhizal roots under different soil phosphate conditions [J]. Mol Plant Microbe Interact, 1993, 6(1): 75-83. |
| [32] | Przysiecka Ł, Książkiewicz M, Wolko B, et al. Structure, expression profile and phylogenetic inference of chalcone isomerase-like genes from the narrow-leafed lupin (Lupinus angustifolius L.) genome [J]. Front Plant Sci, 2015, 6: 268. |
| [33] | 黎霜. 利用樱桃查尔酮合酶CpCHS1基因创制烟草抗旱种质 [D]. 贵阳: 贵州大学, 2022. |
| Li S. Using cherry chalcone synthase CpCHS1 gene to create drought-resistant tobacco germplasm [D]. Guiyang: Guizhou University, 2022. | |
| [34] | Wang CC, Wu DW, Jiang LY, et al. Multi-omics elucidates difference in accumulation of bioactive constituents in licorice (Glycyrrhiza uralensis) under drought stress [J]. Molecules, 2023, 28(20): 7042. |
| [35] | Negi S, Madari S, Tak H, et al. Studies on the tissue specific nature and stress inducible activation of the CHI-1 gene from banana [J]. Plant Physiol Biochem, 2021, 168: 62-69. |
| [36] | 曾凤, 张淑娟, 李雯. 采前喷洒赤霉素对采后"贵妃"芒果外观品质的影响 [J]. 保鲜与加工, 2017, 17(2): 7-11. |
| Zeng F, Zhang SJ, Li W. Effects of pre-harvest spraying with gibberellic acid on appearance quality of postharvest ‘Guifei’ mango [J]. Storage Process, 2017, 17(2): 7-11. |
| [1] | 徐小萍, 杨成龙, 和兴, 郭文杰, 吴健, 方少忠. 百合LoAPS1克隆及其在休眠解除过程的功能分析[J]. 生物技术通报, 2025, 41(9): 195-206. |
| [2] | 董向向, 缪百灵, 许贺娟, 陈娟娟, 李亮杰, 龚守富, 朱庆松. 森林草莓FveBBX20基因的生物信息学分析及开花调控功能[J]. 生物技术通报, 2025, 41(9): 115-123. |
| [3] | 李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158. |
| [4] | 刘泽洲, 段乃彬, 岳丽昕, 王清华, 姚行浩, 高莉敏, 孔素萍. 大蒜叶片蜡质成分分析及蜡质缺失基因Ggl-1筛选[J]. 生物技术通报, 2025, 41(9): 219-231. |
| [5] | 腊贵晓, 赵玉龙, 代丹丹, 余永亮, 郭红霞, 史贵霞, 贾慧, 杨铁钢. 红花质膜H+-ATPase基因家族成员鉴定及响应低氮低磷胁迫的表达分析[J]. 生物技术通报, 2025, 41(8): 220-233. |
| [6] | 程雪, 付颖, 柴晓娇, 王红艳, 邓欣. 谷子LHC基因家族鉴定及非生物胁迫表达分析[J]. 生物技术通报, 2025, 41(8): 102-114. |
| [7] | 赖诗雨, 梁巧兰, 魏列新, 牛二波, 陈应娥, 周鑫, 杨思正, 王博. NbJAZ3在苜蓿花叶病毒侵染本氏烟过程中的作用[J]. 生物技术通报, 2025, 41(8): 186-196. |
| [8] | 张学琼, 潘素君, 李魏, 戴良英. 植物磷酸盐转运蛋白在胁迫响应中的研究进展[J]. 生物技术通报, 2025, 41(7): 28-36. |
| [9] | 李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138. |
| [10] | 韩燚, 侯昌林, 唐露, 孙璐, 谢晓东, 梁晨, 陈小强. 大麦HvERECTA基因的克隆及功能分析[J]. 生物技术通报, 2025, 41(7): 106-116. |
| [11] | 李霞, 张泽伟, 刘泽军, 王楠, 郭江波, 辛翠花, 张彤, 简磊. 马铃薯转录因子StMYB96的克隆及功能研究[J]. 生物技术通报, 2025, 41(7): 181-192. |
| [12] | 龚钰涵, 陈兰, 尚方慧子, 郝灵颖, 刘硕谦. 茶树TRB基因家族鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(7): 214-225. |
| [13] | 魏雨佳, 李岩, 康语涵, 弓晓楠, 杜敏, 涂岚, 石鹏, 于子涵, 孙彦, 张昆. 白颖苔草CrMYB4基因的克隆和表达分析[J]. 生物技术通报, 2025, 41(7): 248-260. |
| [14] | 安苗苗, 蔺祥淏, 梁瑞娜, 赵国柱. 碳源对产甲烷菌群调控及产甲烷能力的影响[J]. 生物技术通报, 2025, 41(6): 355-366. |
| [15] | 吴浩, 董伟峰, 贺子天, 李艳肖, 谢辉, 孙明哲, 沈阳, 孙晓丽. 水稻BXL基因家族的全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(6): 87-98. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||