








生物技术通报 ›› 2025, Vol. 41 ›› Issue (9): 94-104.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0142
卢瑶1( ), 袁平平2, 金鑫3, 毛向红1, 范向斌1, 白小东1(
), 袁平平2, 金鑫3, 毛向红1, 范向斌1, 白小东1( )
)
收稿日期:2025-02-12
出版日期:2025-09-26
发布日期:2025-09-24
通讯作者:
白小东,男,研究方向 :马铃薯育种与栽培技术;E-mail: bxd5561@126.com作者简介:卢瑶,女,研究方向 :马铃薯遗传育种;E-mail: luyao041800@163.com
基金资助:
LU Yao1( ), YUAN Ping-ping2, JIN Xin3, MAO Xiang-hong1, FAN Xiang-bin1, BAI Xiao-dong1(
), YUAN Ping-ping2, JIN Xin3, MAO Xiang-hong1, FAN Xiang-bin1, BAI Xiao-dong1( )
)
Received:2025-02-12
Published:2025-09-26
Online:2025-09-24
摘要:
目的 从分子水平分析马铃薯野生种、地方种的遗传多样性并建立马铃薯种质资源的指纹图谱库。 方法 利用简单串接重复序列(simple sequence repeat, SSR)分子标记技术,对国际马铃薯中心亚太中心保存的73份马铃薯野生种、地方种的遗传多样性进行研究。 结果 平均等位基因数(Na)为1.843 7,Nei’s平均基因多样性指数为0.065 9,平均Shannon信息指数为0.128 6,多态性信息含量PIC平均值为0.848 4,表明供试的73份马铃薯种质资源多态性高但遗传差异小。Structure群体结构将73份马铃薯种质分为7个类群,UPGMA聚类分析分为4个类群。 结论 从14对引物中筛选出可完全区分73份种质的5对核心引物,构建了73份马铃薯野生种和地方种的指纹图谱,为后续马铃薯种质鉴定和筛选体系的研究提供了理论基础和技术支撑。
卢瑶, 袁平平, 金鑫, 毛向红, 范向斌, 白小东. 基于SSR标记的马铃薯野生种和地方种遗传多样性分析和指纹图谱构建[J]. 生物技术通报, 2025, 41(9): 94-104.
LU Yao, YUAN Ping-ping, JIN Xin, MAO Xiang-hong, FAN Xiang-bin, BAI Xiao-dong. Genetic Diversity Analysis and Fingerprinting of Wild Potato and Landraces Based on SSR Markers[J]. Biotechnology Bulletin, 2025, 41(9): 94-104.
编号 No. | 引物名称 Name | 引物序列 Primer sequence (5′-3′) | 染色体 Chromosome | 片段大小 Fragment (bp) | 荧光 Tag |
|---|---|---|---|---|---|
| P1 | PM0936 | TGCACTTTCTTTGTCACCATTC TGACCTACTGTATGCTTTGGAA | Ⅸ | 102-151 | FAM |
| P2 | PM0642 | TCCATATGCAAGTGGCAAAA CACGTTGGATCCAGGAGGTA | Ⅹ | 112-163 | FAM |
| P3 | PM0938 | AGAAATTGGCAGAGCATTTAGCTG GGATTAGACAAACCTTCTTTTCCACA | Ⅲ | 137-177 | FAM |
| P4 | PM0355 | CCATCATGAGCCTTGCTTACATTT CAACAAGGCACAAAGCTAACAGAA | Ⅷ | 172-235 | FAM |
| P5 | STM1052 | CGTTAAGGAGGAGGAGGAAAA CCAAATAACGTGTTGAGCCC | Ⅳ | 199-262 | FAM |
| P6 | STI032 | TAGGGATGAGCTTCAACTCG CGGTTCTGACTTGGACTAATGA | Ⅶ | 100-127 | FAM |
| P7 | STI033 | CAATTTCGTTTTTTCATGTGACAC ATGGCGTAATTTGATTTAATACGTAA | Ⅸ | 109-133 | FAM |
| P8 | STG0016 | ATCCTTCTATGGCGTTCCAA ATTGCTCAAGAATGGCCAC | Ⅶ | 116-156 | FAM |
| P9 | STM0019a | TGAGGGTTTTCAGAAAGGGA CATCCTTGCAACAACCTCCT | Ⅶ | 133-233 | FAM |
| P10 | STM3012 | TATGGAAATTCCGGTGATGG GACGGTGACAAAGAGGAAGG | Ⅺ | 145-267 | FAM |
| P11 | STI0012 | AATAGGTGTACTGACTCTCAATG TTGAAGTAAAAGTCCTAGTATGTG | Ⅵ | 152-199 | FAM |
| P12 | STM2022 | TGATTCTCTTGCCTACTGTAATCG CAAAGTGGTGTGAAGCTGTGA | Ⅷ | 162-341 | FAM |
| P13 | STM1049 | TCTCCCCATCTTAATGTTTC CAACACAGCATACAGATCATC | Ⅲ | 168-197 | FAM |
| P14 | SSR0387 | TTGATGAAAGGAATGCAGCTTGTG ACGTTAAAGAAGTGAGAGTACGAC | Ⅴ | 141-187 | FAM |
表1 14对SSR引物信息
Table 1 Information on 14 pairs SSR primers
编号 No. | 引物名称 Name | 引物序列 Primer sequence (5′-3′) | 染色体 Chromosome | 片段大小 Fragment (bp) | 荧光 Tag |
|---|---|---|---|---|---|
| P1 | PM0936 | TGCACTTTCTTTGTCACCATTC TGACCTACTGTATGCTTTGGAA | Ⅸ | 102-151 | FAM |
| P2 | PM0642 | TCCATATGCAAGTGGCAAAA CACGTTGGATCCAGGAGGTA | Ⅹ | 112-163 | FAM |
| P3 | PM0938 | AGAAATTGGCAGAGCATTTAGCTG GGATTAGACAAACCTTCTTTTCCACA | Ⅲ | 137-177 | FAM |
| P4 | PM0355 | CCATCATGAGCCTTGCTTACATTT CAACAAGGCACAAAGCTAACAGAA | Ⅷ | 172-235 | FAM |
| P5 | STM1052 | CGTTAAGGAGGAGGAGGAAAA CCAAATAACGTGTTGAGCCC | Ⅳ | 199-262 | FAM |
| P6 | STI032 | TAGGGATGAGCTTCAACTCG CGGTTCTGACTTGGACTAATGA | Ⅶ | 100-127 | FAM |
| P7 | STI033 | CAATTTCGTTTTTTCATGTGACAC ATGGCGTAATTTGATTTAATACGTAA | Ⅸ | 109-133 | FAM |
| P8 | STG0016 | ATCCTTCTATGGCGTTCCAA ATTGCTCAAGAATGGCCAC | Ⅶ | 116-156 | FAM |
| P9 | STM0019a | TGAGGGTTTTCAGAAAGGGA CATCCTTGCAACAACCTCCT | Ⅶ | 133-233 | FAM |
| P10 | STM3012 | TATGGAAATTCCGGTGATGG GACGGTGACAAAGAGGAAGG | Ⅺ | 145-267 | FAM |
| P11 | STI0012 | AATAGGTGTACTGACTCTCAATG TTGAAGTAAAAGTCCTAGTATGTG | Ⅵ | 152-199 | FAM |
| P12 | STM2022 | TGATTCTCTTGCCTACTGTAATCG CAAAGTGGTGTGAAGCTGTGA | Ⅷ | 162-341 | FAM |
| P13 | STM1049 | TCTCCCCATCTTAATGTTTC CAACACAGCATACAGATCATC | Ⅲ | 168-197 | FAM |
| P14 | SSR0387 | TTGATGAAAGGAATGCAGCTTGTG ACGTTAAAGAAGTGAGAGTACGAC | Ⅴ | 141-187 | FAM |
序号 Code | 引物 Primer | Na* | Ne* | H* | I* | 多态位点 Polymorphic alleles | 多态百分率 Percentage of polymorphic bands (%) | PIC |
|---|---|---|---|---|---|---|---|---|
| P1 | PM0936 | 1.952 4 | 1.085 3 | 0.074 2 | 0.152 3 | 20 | 95.24 | 0.917 7 |
| P2 | PM0642 | 1.923 1 | 1.049 9 | 0.042 7 | 0.094 8 | 36 | 92.31 | 0.917 2 |
| P3 | PM0938 | 1.963 | 1.062 3 | 0.054 3 | 0.116 6 | 26 | 96.3 | 0.914 5 |
| P4 | PM0355 | 1.731 7 | 1.033 8 | 0.031 | 0.071 9 | 30 | 73.17 | 0.936 5 |
| P5 | STM1052 | 1.666 7 | 1.042 8 | 0.036 6 | 0.078 4 | 22 | 66.67 | 0.884 9 |
| P6 | STI032 | 1.8 | 1.159 7 | 0.114 8 | 0.200 8 | 8 | 80 | 0.758 5 |
| P7 | STI033 | 1.818 2 | 1.147 8 | 0.102 | 0.183 | 9 | 81.82 | 0.723 9 |
| P8 | STG0016 | 1.866 7 | 1.117 3 | 0.093 1 | 0.176 4 | 13 | 86.67 | 0.856 5 |
| P9 | STM0019a | 1.808 5 | 1.033 7 | 0.030 3 | 0.071 | 38 | 80.85 | 0.931 9 |
| P10 | STM3012 | 1.889 | 1.061 8 | 0.041 4 | 0.082 7 | 24 | 88.89 | 0.683 2 |
| P11 | STI0012 | 2.0 | 1.089 9 | 0.070 2 | 0.135 3 | 16 | 100 | 0.809 9 |
| P12 | STM2022 | 1.575 8 | 1.075 9 | 0.054 8 | 0.102 | 19 | 57.58 | 0.850 2 |
| P13 | STM1049 | 1.916 7 | 1.128 9 | 0.096 5 | 0.174 9 | 11 | 91.67 | 0.787 1 |
| P14 | SSR0387 | 1.9 | 1.095 1 | 0.080 5 | 0.159 8 | 18 | 90 | 0.906 4 |
| 均值 Mean | 1.843 7 | 1.084 6 | 0.065 9 | 0.128 6 | 20.71 | 84.37 | 0.848 4 | |
表2 14对SSR引物在73份马铃薯种质资源中的多态性检测
Table 2 Polymorphic detection of 14 SSR primers in 73 potato germplasm resources
序号 Code | 引物 Primer | Na* | Ne* | H* | I* | 多态位点 Polymorphic alleles | 多态百分率 Percentage of polymorphic bands (%) | PIC |
|---|---|---|---|---|---|---|---|---|
| P1 | PM0936 | 1.952 4 | 1.085 3 | 0.074 2 | 0.152 3 | 20 | 95.24 | 0.917 7 |
| P2 | PM0642 | 1.923 1 | 1.049 9 | 0.042 7 | 0.094 8 | 36 | 92.31 | 0.917 2 |
| P3 | PM0938 | 1.963 | 1.062 3 | 0.054 3 | 0.116 6 | 26 | 96.3 | 0.914 5 |
| P4 | PM0355 | 1.731 7 | 1.033 8 | 0.031 | 0.071 9 | 30 | 73.17 | 0.936 5 |
| P5 | STM1052 | 1.666 7 | 1.042 8 | 0.036 6 | 0.078 4 | 22 | 66.67 | 0.884 9 |
| P6 | STI032 | 1.8 | 1.159 7 | 0.114 8 | 0.200 8 | 8 | 80 | 0.758 5 |
| P7 | STI033 | 1.818 2 | 1.147 8 | 0.102 | 0.183 | 9 | 81.82 | 0.723 9 |
| P8 | STG0016 | 1.866 7 | 1.117 3 | 0.093 1 | 0.176 4 | 13 | 86.67 | 0.856 5 |
| P9 | STM0019a | 1.808 5 | 1.033 7 | 0.030 3 | 0.071 | 38 | 80.85 | 0.931 9 |
| P10 | STM3012 | 1.889 | 1.061 8 | 0.041 4 | 0.082 7 | 24 | 88.89 | 0.683 2 |
| P11 | STI0012 | 2.0 | 1.089 9 | 0.070 2 | 0.135 3 | 16 | 100 | 0.809 9 |
| P12 | STM2022 | 1.575 8 | 1.075 9 | 0.054 8 | 0.102 | 19 | 57.58 | 0.850 2 |
| P13 | STM1049 | 1.916 7 | 1.128 9 | 0.096 5 | 0.174 9 | 11 | 91.67 | 0.787 1 |
| P14 | SSR0387 | 1.9 | 1.095 1 | 0.080 5 | 0.159 8 | 18 | 90 | 0.906 4 |
| 均值 Mean | 1.843 7 | 1.084 6 | 0.065 9 | 0.128 6 | 20.71 | 84.37 | 0.848 4 | |
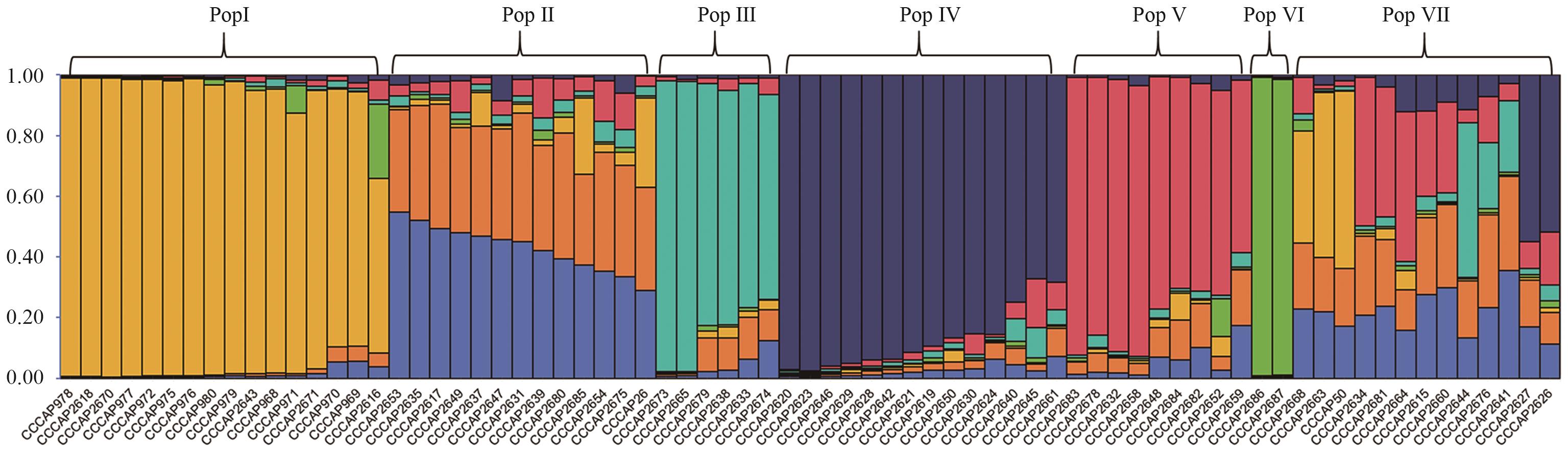
图2 K值为7的73份马铃薯种质资源群体结构每1竖条代表1个样本,且不同的颜色代表来源于不同的种
Fig. 2 Population structure of 73 potato germplasm resources on K value of 7Each vertical bar refers to a sample, and different colors refer to different pedigrees
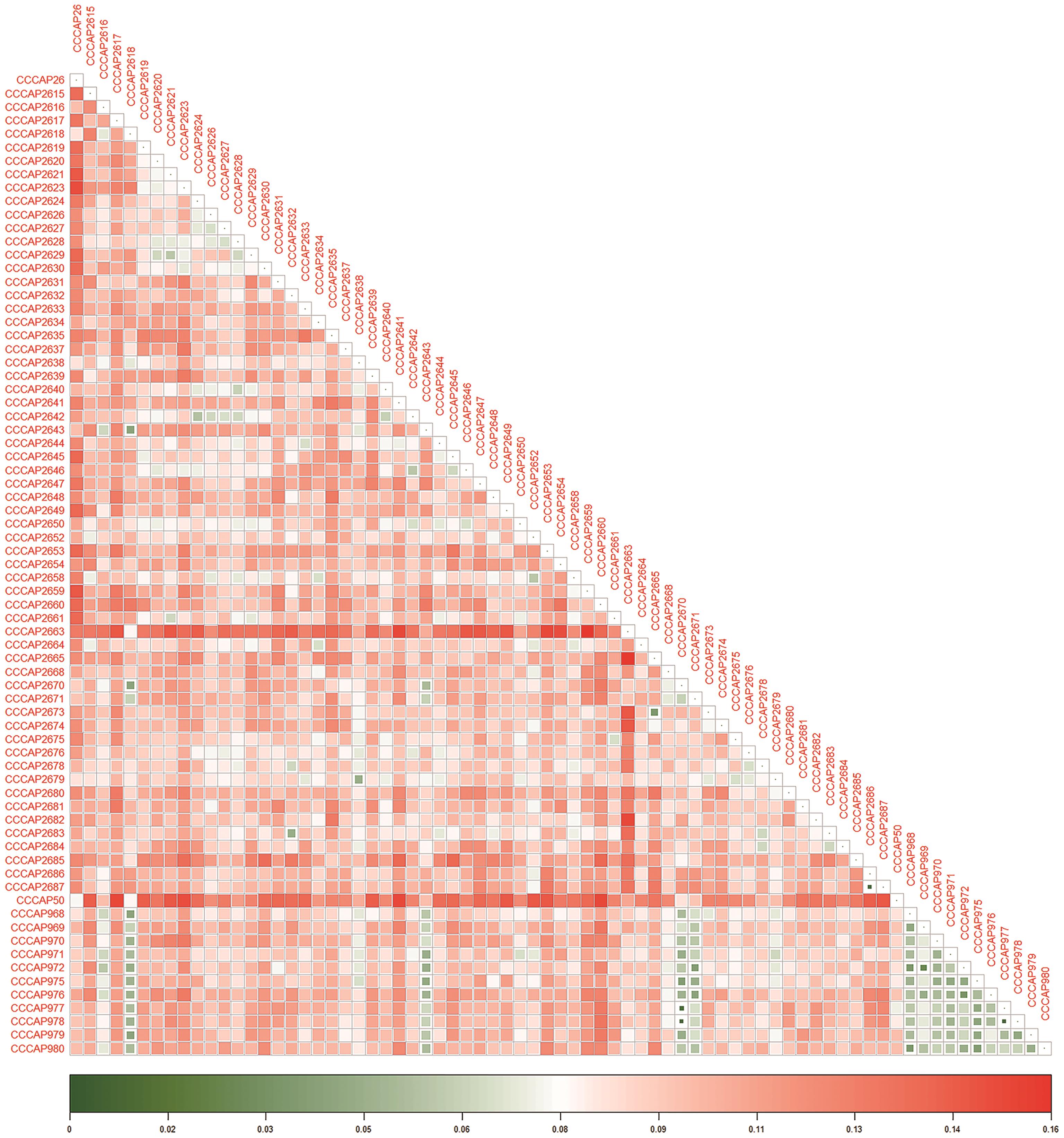
图3 73份马铃薯种质资源遗传距离热图采用颜色梯度表示遗传距离,绿色至红色的渐变对应遗传距离0-0.157的连续变化,其中深绿色指示遗传距离趋近于0,而深红色则反映遗传距离的增加
Fig. 3 Heat map of genetic distances of 73 potato germplasm resourcesA color gradient was employed to indicate genetic distance, with a transition from green to red corresponding to a continuous range of genetic distances from 0 to 0.157. Deep green indicates a genetic distance approaching 0, while deep red reflects an increase in genetic distance

图5 73份马铃薯样品的SSR标记主坐标分析pop 1是地方种,pop 2是品种,pop 3是野生种
Fig. 5 Principal coordinates analysis of SSR markers in 73 potato samplespop 1 refers to landraces, pop 2 refers to variety, pop 3 refers to wild potatoes
编号 No. | CCCAP编号 GCD code | SSR指纹 SSR fingerprinting | 编号 No. | CCCAP编号 GCD code | SSR指纹 SSR fingerprinting |
|---|---|---|---|---|---|
| 1 | CCCAP26 | 17C3639478 | 38 | CCCAP2642 | 7HGI589 |
| 2 | CCCAP50 | 178469JN34638E | 39 | CCCAP2643 | 84639BH78 |
| 3 | CCCAP968 | 869988 | 40 | CCCAP2644 | 2F369 |
| 4 | CCCAP969 | 8JL988 | 41 | CCCAP2645 | 368A34BD659 |
| 5 | CCCAP970 | 8I563988 | 42 | CCCAP2646 | 37A4D558 |
| 6 | CCCAP971 | 8463778 | 43 | CCCAP2647 | KNO46DF8A1 |
| 7 | CCCAP972 | 86988 | 44 | CCCAP2648 | 123B671 |
| 8 | CCCAP975 | 86396738 | 45 | CCCAP2649 | 318G81 |
| 9 | CCCAP976 | 584I3988 | 46 | CCCAP2650 | 8E3K98 |
| 10 | CCCAP977 | 84I9JN738 | 47 | CCCAP2652 | 6963B77A |
| 11 | CCCAP978 | 84I9738 | 48 | CCCAP2653 | 145I9H561 |
| 12 | CCCAP979 | 8F69738 | 49 | CCCAP2654 | D5919H5E |
| 13 | CCCAP980 | 386397838 | 50 | CCCAP2658 | 9A3686 |
| 14 | CCCAP2615 | 69FGI3693 | 51 | CCCAP2659 | EJ7161 |
| 15 | CCCAP2616 | 135469717 | 52 | CCCAP2660 | BC7E3J61 |
| 16 | CCCAP2617 | 5L4Q39582 | 53 | CCCAP2661 | 3E3J6A |
| 17 | CCCAP2618 | 8F469H88 | 54 | CCCAP2663 | 8CF6E39JN683689 |
| 18 | CCCAP2619 | 46D34569 | 55 | CCCAP2664 | 8K53561 |
| 19 | CCCAP2620 | 46CEJ568D | 56 | CCCAP2665 | 494D48 |
| 20 | CCCAP2621 | 3CFJ56AC | 57 | CCCAP2668 | H53837 |
| 21 | CCCAP2623 | 3CJ34JK536 | 58 | CCCAP2670 | 84I9H738 |
| 22 | CCCAP2624 | 57FI55 | 59 | CCCAP2671 | 7FG39838 |
| 23 | CCCAP2626 | BHI656 | 60 | CCCAP2673 | 4679448 |
| 24 | CCCAP2627 | 9FR1I61 | 61 | CCCAP2674 | 46673 |
| 25 | CCCAP2628 | 59CJ362 | 62 | CCCAP2675 | 3853568 |
| 26 | CCCAP2629 | 35CE3J568E | 63 | CCCAP2676 | 7A8969 |
| 27 | CCCAP2630 | 9AB34JK566E | 64 | CCCAP2678 | 6A8962 |
| 28 | CCCAP2631 | 66E982 | 65 | CCCAP2679 | C639468 |
| 29 | CCCAP2632 | 697M3B61 | 66 | CCCAP2680 | 6B6G39H687E |
| 30 | CCCAP2633 | CK3739 | 67 | CCCAP2681 | 7C5H3B66A |
| 31 | CCCAP2634 | 715361 | 68 | CCCAP2682 | 7583B51 |
| 32 | CCCAP2635 | 39JO59EH68 | 69 | CCCAP2683 | 67G3B61 |
| 33 | CCCAP2637 | 749H6A1 | 70 | CCCAP2684 | 6A539BH626 |
| 34 | CCCAP2638 | 4D639689 | 71 | CCCAP2685 | EF459EH88A |
| 35 | CCCAP2639 | 9B693961 | 72 | CCCAP2686 | 3E623AB67 |
| 36 | CCCAP2640 | 7CF13IJ658 | 73 | CCCAP2687 | 3E623AB657 |
| 37 | CCCAP2641 | 17I359B |
表3 73份马铃薯种质资源指纹图谱
Table 3 Fingerprints of 73 potato germplasm resources
编号 No. | CCCAP编号 GCD code | SSR指纹 SSR fingerprinting | 编号 No. | CCCAP编号 GCD code | SSR指纹 SSR fingerprinting |
|---|---|---|---|---|---|
| 1 | CCCAP26 | 17C3639478 | 38 | CCCAP2642 | 7HGI589 |
| 2 | CCCAP50 | 178469JN34638E | 39 | CCCAP2643 | 84639BH78 |
| 3 | CCCAP968 | 869988 | 40 | CCCAP2644 | 2F369 |
| 4 | CCCAP969 | 8JL988 | 41 | CCCAP2645 | 368A34BD659 |
| 5 | CCCAP970 | 8I563988 | 42 | CCCAP2646 | 37A4D558 |
| 6 | CCCAP971 | 8463778 | 43 | CCCAP2647 | KNO46DF8A1 |
| 7 | CCCAP972 | 86988 | 44 | CCCAP2648 | 123B671 |
| 8 | CCCAP975 | 86396738 | 45 | CCCAP2649 | 318G81 |
| 9 | CCCAP976 | 584I3988 | 46 | CCCAP2650 | 8E3K98 |
| 10 | CCCAP977 | 84I9JN738 | 47 | CCCAP2652 | 6963B77A |
| 11 | CCCAP978 | 84I9738 | 48 | CCCAP2653 | 145I9H561 |
| 12 | CCCAP979 | 8F69738 | 49 | CCCAP2654 | D5919H5E |
| 13 | CCCAP980 | 386397838 | 50 | CCCAP2658 | 9A3686 |
| 14 | CCCAP2615 | 69FGI3693 | 51 | CCCAP2659 | EJ7161 |
| 15 | CCCAP2616 | 135469717 | 52 | CCCAP2660 | BC7E3J61 |
| 16 | CCCAP2617 | 5L4Q39582 | 53 | CCCAP2661 | 3E3J6A |
| 17 | CCCAP2618 | 8F469H88 | 54 | CCCAP2663 | 8CF6E39JN683689 |
| 18 | CCCAP2619 | 46D34569 | 55 | CCCAP2664 | 8K53561 |
| 19 | CCCAP2620 | 46CEJ568D | 56 | CCCAP2665 | 494D48 |
| 20 | CCCAP2621 | 3CFJ56AC | 57 | CCCAP2668 | H53837 |
| 21 | CCCAP2623 | 3CJ34JK536 | 58 | CCCAP2670 | 84I9H738 |
| 22 | CCCAP2624 | 57FI55 | 59 | CCCAP2671 | 7FG39838 |
| 23 | CCCAP2626 | BHI656 | 60 | CCCAP2673 | 4679448 |
| 24 | CCCAP2627 | 9FR1I61 | 61 | CCCAP2674 | 46673 |
| 25 | CCCAP2628 | 59CJ362 | 62 | CCCAP2675 | 3853568 |
| 26 | CCCAP2629 | 35CE3J568E | 63 | CCCAP2676 | 7A8969 |
| 27 | CCCAP2630 | 9AB34JK566E | 64 | CCCAP2678 | 6A8962 |
| 28 | CCCAP2631 | 66E982 | 65 | CCCAP2679 | C639468 |
| 29 | CCCAP2632 | 697M3B61 | 66 | CCCAP2680 | 6B6G39H687E |
| 30 | CCCAP2633 | CK3739 | 67 | CCCAP2681 | 7C5H3B66A |
| 31 | CCCAP2634 | 715361 | 68 | CCCAP2682 | 7583B51 |
| 32 | CCCAP2635 | 39JO59EH68 | 69 | CCCAP2683 | 67G3B61 |
| 33 | CCCAP2637 | 749H6A1 | 70 | CCCAP2684 | 6A539BH626 |
| 34 | CCCAP2638 | 4D639689 | 71 | CCCAP2685 | EF459EH88A |
| 35 | CCCAP2639 | 9B693961 | 72 | CCCAP2686 | 3E623AB67 |
| 36 | CCCAP2640 | 7CF13IJ658 | 73 | CCCAP2687 | 3E623AB657 |
| 37 | CCCAP2641 | 17I359B |
| [1] | Food and Agriculture Organization of the United Nations. https://www.fao.org/faostat/en/#data/QCL [DB/OL]. 2022. |
| [2] | Devaux A, Goffart JP, Kromann P, et al. The potato of the future: opportunities and challenges in sustainable agri-food systems [J]. Potato Res, 2021, 64(4): 681-720. |
| [3] | 宋波涛, 司怀军, 聂碧华. 马铃薯种质资源利用与改良 [M]. 呼和浩特: 内蒙古人民出版社, 2024. |
| Song BT, Si HJ, Nie BH. Utilization and improvement of potato germplasm resources [M]. Hohhot: China: Inner Mongolian People’s Publishing House, 2024. | |
| [4] | Spooner DM, Ghislain M, Simon R, et al. Systematics, diversity, genetics, and evolution of wild and cultivated potatoes [J]. Bot Rev, 2014, 80(4): 283-383. |
| [5] | Jenderek MM, Ambruzs BD, Tanner JD, et al. High regrowth of potato crop wild relative genotypes after cryogenic storage [J]. Cryobiology, 2023, 111: 84-88. |
| [6] | 段绍光, 金黎平, 李广存, 等. 马铃薯品种遗传多样性分析 [J]. 作物学报, 2017, 43 (5): 718-729. |
| Duan SG, Jin LP, Li GC, et al. Analysis of genetic diversity of potato varieties [J]. Acta Agronomica Sinica, 2017, 43 (5): 718-729. | |
| [7] | Ali J, Sobhy IS, Bruce TJ. Wild potato ancestors as potential sources of resistance to the aphid Myzus persicae [J]. Pest Manag Sci, 2022, 78(9): 3931-3938. |
| [8] | Gorzolka K, Perino EHB, Lederer S, et al. Lysophosphatidylcholine 17: 1 from the leaf surface of the wild potato species Solanum bulbocastanum inhibits Phytophthora infestans [J]. J Agric Food Chem, 2021, 69(20): 5607-5617. |
| [9] | Xu Y, Gray SM. Aphids and their transmitted potato viruses: a continuous challenges in potato crops [J]. J Integr Agric, 2020, 19(2): 367-375. |
| [10] | Karki HS, Jansky SH, Halterman DA. Screening of wild potatoes identifies new sources of late blight resistance [J]. Plant Dis, 2021, 105(2): 368-376. |
| [11] | Hawkes JG. The potato: evolution, biodiversity and genetic resources [M]. Washington, D.C.: Smithsonian Institution Press, 1990. |
| [12] | Spooner DM, Rodríguez F, Polgár Z, et al. Genomic origins of potato polyploids: GBSSI gene sequencing data [J]. Crop Sci, 2008, 48(S1): 27-36. |
| [13] | Li YP, Colleoni C, Zhang JJ, et al. Genomic analyses yield markers for identifying agronomically important genes in potato [J]. Mol Plant, 2018, 11(3): 473-484. |
| [14] | 宋晓燕, 张春芝, 李颖, 等. 二倍体马铃薯栽培品种遗传多样性的SSR标记分析 [J]. 园艺学报, 2016, 43 (11): 2266-2276. |
| Song XY, Zhang CZ, Li Y, et al. SSR marker analysis of genetic diversity in diploid potato cultivars [J]. Acta Horticulturae Sinica, 2016, 43 (11): 2266-2276. | |
| [15] | 曹越, 张立媛, 辛旭霞, 等. 基于荧光SSR的宁夏糜子DNA分子身份证的构建 [J]. 作物学报, 2024, 50 (11): 2699-2711. |
| Cao Y, Zhang LY, Xin XX, et al. Construction of DNA molecular identity card of proso millet in Ningxia based on fluorescent SSR [J]. Acta Agronomica Sinica, 2024, 50 (11): 2699-2711. | |
| [16] | 王秀军, 赵彦贝, 王静, 等. 基于SSR分子标记的175份蜡梅种质遗传多样性分析和指纹图谱构建 [J]. 生物工程学报, 2024, 40(1): 252-268. |
| Wang XJ, Zhao YB, Wang J, et al. Genetic diversity analysis and fingerprinting of 175 Chimonanthus praecox germplasm based on SSR molecular marker [J]. Chin J Biotechnol, 2024, 40(1): 252-268. | |
| [17] | 韩嘉琪, 王海平, 宋江萍, 等. 基于SSR标记的百合资源遗传背景分析 [J]. 生物工程学报, 2024, 40(4): 1211-1224. |
| Han JQ, Wang HP, Song JP, et al. Genetic background of lily germplasm resources based on SSR markers [J]. China Ind Econ, 2024, 40(4): 1211-1224. | |
| [18] | 王晓映, 张方玉, 万星, 等. 基于分子标记和表型性状的水稻地方品种遗传多样性研究 [J]. 植物遗传资源学报, 2023, 24 (3): 636-647. |
| Wang XY, Zhang FY, Wan X, et al. Genetic diversity of local varieties of rice based on molecular markers and phenotypic traits [J]. Journal of Plant Genetic Resources, 2023, 24(3):636-647. | |
| [19] | Tillault AS, Yevtushenko DP. Simple sequence repeat analysis of new potato varieties developed in Alberta, Canada [J]. Plant Direct, 2019, 3(6): e00140. |
| [20] | 孙小琼, 王芳, 王崇, 等. 马铃薯品种(系)资源的SSR遗传多样性分析及指纹图谱构建 [J/OL]. 分子植物育种, 2022: 1-27. |
| Sun XQ, Wang F, Wang C, et al. SSR genetic diversity analysis and fingerprinting of potato variety (line) resources [J/OL]. Mol Plant Breed, 2022: 1-27. | |
| [21] | Achakkagari SR, Bozan I, Camargo-Tavares JC, et al. The phased Solanum okadae genome and Petota pangenome analysis of 23 other potato wild relatives and hybrids [J]. Sci Data, 2024, 11(1): 454. |
| [22] | Hosaka AJ, Sanetomo R, Hosaka K. A de novo genome assembly of Solanum bulbocastanum Dun., a Mexican diploid species reproductively isolated from the A-genome species, including cultivated potatoes [J]. G3, 2024, 14(6): jkae080. |
| [23] | Hardigan MA, Bamberg J, Robin Buell C, et al. Taxonomy and genetic differentiation among wild and cultivated germplasm of Solanum sect. petota [J]. Plant Genome, 2015, 8(1): eplantgenome2014.06.0025. |
| [24] | 毛向红, 卢瑶, 范向斌, 等. 基于SSR荧光标记毛细管电泳的马铃薯品种遗传多样性分析及分子身份证构建 [J]. 生物技术通报, 2024, (9): 131-140. |
| Mao XH, Lu Y, Fan XB, et al. Genetic diversity analysis of potato varieties based on SSR fluorescent marker capillary electrophoresis and construction of molecular identity card [J]. China Ind Econ, 2024, (9): 131-140. | |
| [25] | 毛立彦, 李慧敏, 龙凌云, 等. 基于SSR分子标记的睡莲遗传多样性分析 [J]. 南京林业大学学报: 自然科学版, 2024, 48(5), 57-68. |
| Mao LY, Li HM, Long LY, et al. Analyzing the genetic diversity of Nymphaea spp. based on SSR markers [J]. Journal of Nanjing Forestry University Natural Sciences Edition, 2024, 48(5), 57-68. | |
| [26] | 梁燕, 韩传明, 孙超, 等. 基于SSR标记的核桃种质资源遗传多样性与遗传结构分析 [J]. 北方园艺, 2022(9): 47-54. |
| Liang Y, Han CM, Sun C, et al. Genetic diversity and genetic structure analysis of walnut germplasm resources based on SSR markers [J]. North Hortic, 2022(9): 47-54. | |
| [27] | Tiwari JK, Ali N, Devi S, et al. Analysis of allelic variation in wild potato (Solanum) species by simple sequence repeat (SSR) markers [J]. 3 Biotech, 2019, 9(7): 262. |
| [28] | Hu J, Mei M, Jin F, et al. Phenotypic variability and genetic diversity analysis of cultivated potatoes in China [J]. Front Plant Sci, 2022, 13: 954162. |
| [29] | 王衍莉, 杨义明, 范书田, 等. 基于SSR分子标记的73份山葡萄及杂交后代的遗传多样性分析 [J].生物技术通报, 2021, 37(1): 189-197. |
| Wang YL, Yang YM, Fan ST, et al. Genetic diversity analysis of 73 Vitis amurensis and its hybrids offsprings based on SSR molecular markers [J]. Biotechnology Bulletin, 2021, 37(1):189-197. | |
| [30] | 闫国跃, 杨帆, 白燕远, 等. 69份苦玄参种质SSR遗传多样性及品质性状相关标记分析 [J]. 中国实验方剂学杂志, 2020, 26(4): 174-184. |
| Yan GY, Yang F, Bai YY, et al. SSR-based analysis of genetic diversity and quality trait-related markers of 69 Picria felterrae germplasm samples [J]. Chin J Exp Tradit Med Formulae, 2020, 26(4): 174-184. | |
| [31] | 赵成日. 中韩野生软枣猕猴桃种质资源遗传多样性分析 [J]. 果树学报, 2018, 35(9): 1043-1051. |
| Zhao CR. The genetic diversity of wild Actinidia arguta germplasm resources from China and South Korea [J]. J Fruit Sci, 2018, 35(9): 1043-1051. | |
| [32] | Wang Y, Rashid MAR, Li XP, et al. Collection and evaluation of genetic diversity and population structure of potato landraces and varieties in China [J]. Front Plant Sci, 2019, 10: 139. |
| [33] | 刘有春, 刘成, 杨艳敏, 等. 基于EST-SSR标记的越橘栽培种和几个中国野生种的遗传结构分析 [J]. 果树学报, 2017, 34(8): 956-967. |
| Liu YC, Liu C, Yang YM, et al. Genetic structure analysis of the cultivated blueberry (Vaccinium spp.) species and wild species in China based on EST-SSR markers [J]. J Fruit Sci, 2017, 34(8): 956-967. | |
| [34] | 王琰琰, 王俊, 刘国祥, 等. 基于SSR标记的雪茄烟种质资源指纹图谱库的构建及遗传多样性分析 [J]. 作物学报, 2021, 47(7): 1259-1274. |
| Wang YY, Wang J, Liu GX, et al. Construction of SSR fingerprint database and genetic diversity analysis of cigar germplasm resources [J]. Acta Agron Sin, 2021, 47(7): 1259-1274. | |
| [35] | 张晓煜, 王仕鹏, 曹昆山, 等. 基于表型性状与SSR标记的马铃薯种质资源遗传多样性研究 [J]. 西北农业学报, 2024, 33(8):1436-1447. |
| Zhang XY, Wang SP, Cao KS, et al. Genetic diversity of potato germplasm resources based on phenotypic traits and SSR markers [J]. China Ind Econ, 2024, 33(8):1436-1447. | |
| [36] | 孙泽硕, 蒋冬月, 柳新红, 等. 基于SSR标记的42份樱花品种的聚类分析及DNA指纹图谱构建 [J]. 园艺学报, 2023, 50(3): 657-668. |
| Sun ZS, Jiang DY, Liu XH, et al. Cluster analysis and construction of DNA fingerprinting of 42 oriental cultivars of flowering cherry based on SSR markers [J]. Acta Hortic Sin, 2023, 50(3): 657-668. | |
| [37] | 李佩华, 张伦, 罗杰,等. 四川山区63份马铃薯亲本材料的SSR指纹图谱构建及遗传多样性分析 [J]. 四川农业大学学报, 2024, 42(4): 792-799. |
| Li PH, Zhang L, Luo J, et al. Construction of SSR fingerprints and genetic diversity analysis of 63 potato materials in mountainous regions of sichuan province [J]. Journal of Sichuan Agricultural University, 2024,42(4):792-799. | |
| [38] | 王丹, 张志成, 韩海霞, 等. 基于SSR技术的马铃薯种质资源遗传多样性分析 [J]. 作物杂志, 2025, 4: 29-40. |
| Wang D, Zhang ZC, Han HX, et al. Genetic diversity analysis of potato germplasm resources based on SSR technology [J]. Crops, 2025, 4: 29-40. | |
| [39] | 刘文林, 张举梅, 盛万民, 等. 52份俄罗斯引进马铃薯种质资源的遗传多样性与分子身份证构建 [J]. 分子植物育种, 2016, 14(1): 251-258. |
| Liu WL, Zhang JM, Sheng WM, et al. Genetic diversity analysis and molecular identities establishment of 52 Russian potato varieties [J]. Mol Plant Breed, 2016, 14(1): 251-258. |
| [1] | 巩慧玲, 邢玉洁, 马俊贤, 蔡霞, 冯再平. 马铃薯LAC基因家族的鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2025, 41(9): 82-93. |
| [2] | 裴红霞, 汪露瑶, 李生梅, 高晶霞. 基于SCoT、SRAP和SSR分子标记的220份辣椒种质资源遗传多样性分析[J]. 生物技术通报, 2025, 41(8): 165-174. |
| [3] | 段敏杰, 李怡斐, 王春萍, 黄任中, 黄启中, 张世才. 辣椒果实颜色性状与SSR分子标记的关联分析及指纹图谱构建[J]. 生物技术通报, 2025, 41(7): 81-94. |
| [4] | 黄丹丹, 吴云翼, 邹建华, 俞婷, 朱炎辉, 杨梅宏, 董文丽, 高冬丽. 马铃薯StPTST2a基因的克隆及互作分析[J]. 生物技术通报, 2025, 41(7): 172-180. |
| [5] | 李霞, 张泽伟, 刘泽军, 王楠, 郭江波, 辛翠花, 张彤, 简磊. 马铃薯转录因子StMYB96的克隆及功能研究[J]. 生物技术通报, 2025, 41(7): 181-192. |
| [6] | 罗稷林, 栗锦烨, 贾玉鑫. 马铃薯中重力响应调节基因鉴定及功能分析[J]. 生物技术通报, 2025, 41(6): 109-118. |
| [7] | 许慧珍, SHANTWANA Ghimire, RAJU Kharel, 岳云, 司怀军, 唐勋. 马铃薯SUMO E3连接酶基因家族分析及StSIZ1基因的克隆与表达模式分析[J]. 生物技术通报, 2025, 41(6): 119-129. |
| [8] | 段永红, 杨欣, 于冠群, 夏俊俊, 宋陆帅, 白小东, 彭锁堂. 125份马铃薯种质资源遗传多样性及主成分分析[J]. 生物技术通报, 2025, 41(6): 130-143. |
| [9] | 宋慧洋, 苏宝杰, 李京昊, 梅超, 宋倩娜, 崔福柱, 冯瑞云. 马铃薯StAS2-15基因的克隆及盐胁迫下功能分析[J]. 生物技术通报, 2025, 41(5): 119-128. |
| [10] | 文博霖, 万敏, 胡建军, 王克秀, 景晟林, 王心悦, 朱博, 唐铭霞, 李兵, 何卫, 曾子贤. 马铃薯川芋50遗传转化及基因编辑体系的建立[J]. 生物技术通报, 2025, 41(4): 88-97. |
| [11] | 刘涛, 王志淇, 吴文博, 石文婷, 王超楠, 杜崇, 杨中敏. 马铃薯GRAM基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(4): 145-155. |
| [12] | 张益瑄, 马宇, 王童童, 盛苏奥, 宋家凤, 吕钊彦, 朱晓彪, 侯华兰. 马铃薯DIR家族全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(3): 123-136. |
| [13] | 俞婷, 黄丹丹, 朱炎辉, 杨梅宏, 艾菊, 高冬丽. 马铃薯Stpatatin 05基因转录调控因子筛选及互作验证[J]. 生物技术通报, 2025, 41(3): 137-145. |
| [14] | 覃悦, 杨妍, 张磊, 卢丽丽, 李先平, 蒋伟. 二倍体和四倍体马铃薯StGAox基因鉴定与比较分析[J]. 生物技术通报, 2025, 41(3): 146-160. |
| [15] | 于静, 于桂爽, 孙昊杰, 姜春姣, 苑广迪, 杨珍, 王志伟, 王超, 王传堂. 花生种用品质影响因素及相关标记研究[J]. 生物技术通报, 2025, 41(2): 284-294. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||