








生物技术通报 ›› 2025, Vol. 41 ›› Issue (3): 123-136.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0825
• 研究报告 • 上一篇
张益瑄( ), 马宇, 王童童, 盛苏奥, 宋家凤, 吕钊彦, 朱晓彪, 侯华兰(
), 马宇, 王童童, 盛苏奥, 宋家凤, 吕钊彦, 朱晓彪, 侯华兰( )
)
收稿日期:2024-08-25
出版日期:2025-03-26
发布日期:2025-03-20
通讯作者:
侯华兰,女,博士,讲师,研究方向 :蔬菜种质资源与遗传育种;E-mail: hhl@ahau.edu.cn作者简介:张益瑄,女,研究方向 :蔬菜种质资源与遗传育种;E-mail: 15045930755@stu.ahau.edu.cn
基金资助:
ZHANG Yi-xuan( ), MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan(
), MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan( )
)
Received:2024-08-25
Published:2025-03-26
Online:2025-03-20
摘要:
目的 基于最新版马铃薯基因组信息,鉴定DIR基因家族所有成员,并利用生物信息学技术和转录组数据,对DIR基因家族成员系统进化、基因结构、顺式元件及表达模式进行预测及分析,为深入研究DIR基因在马铃薯生长发育及胁迫响应中的功能提供参考。 方法 基于拟南芥DIR基因和Dirigent结构域模型,对马铃薯DIR家族成员进行全基因组鉴定。通过ExPASy、MEME、MCScanX、GSDS、PlantCARE和TBtools等生物信息学工具获取马铃薯DIR基因家族成员的理化特征、染色体定位、共线性、保守基序、基因结构和顺式作用元件等信息。基于转录组数据和定量PCR分析,了解DIR基因在不同组织和胁迫处理下的表达模式,挖掘生长发育及胁迫响应相关StDIR基因。 结果 马铃薯中共鉴定到34个DIR基因,不均匀分布于11条染色体上;StDIR基因分为3个亚组,同一亚组内成员的保守基序和基因结构相对一致,且大部分基因仅含有一个外显子;顺式作用元件分析发现StDIR基因启动子中含有多个激素诱导相关元件和非生物胁迫响应元件;共线性分析显示马铃薯中5个、13个和18个DIR基因分别与水稻、拟南芥和茄子中3个、15个和16个DIR基因存在共线性;表达模式分析发现StDIR基因在叶、根、匍匐茎和愈伤中相对高表达,而在萼片、心皮、雄蕊、花瓣中表达量较低;此外,StDIR基因对盐、干旱、高温和冷胁迫处理具有不同程度的响应,表明其在马铃薯非生物胁迫响应中发挥重要作用。 结论 马铃薯中共鉴定出34个DIR家族成员,分为3个亚组,同一亚组内保守motif和基因结构基本一致。StDIR基因在不同组织间表达模式存在差异,且对干旱和高温胁迫具有较显著响应。
张益瑄, 马宇, 王童童, 盛苏奥, 宋家凤, 吕钊彦, 朱晓彪, 侯华兰. 马铃薯DIR家族全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(3): 123-136.
ZHANG Yi-xuan, MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan. Genome-wide Identification and Expression Profiles of DIR Gene Family in Potato[J]. Biotechnology Bulletin, 2025, 41(3): 123-136.
| 基因名Gene name | 基因ID Gene ID | 蛋白长度Length of protein/aa | 分子量Molecular weight/kD | 等电点pI | 不稳定系数Instability index | 脂肪系数Aliphatic index | 亲水性平均值Grand average of hydropathicity | 信号肽Signal peptide/aa | N-糖基化位点N-Glyc position | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|---|
| StDIR1 | Soltu.DM.01G001610.1 | 192 | 21.26 | 9.76 | 31.2 | 100.47 | 0.167 | 1-24 | 41 | Chloroplast |
| StDIR2 | Soltu.DM.01G012660.1 | 189 | 21.17 | 9.57 | 28 | 86.19 | 0.155 | 1-24 | - | Cell membrane. Chloroplast |
| StDIR3 | Soltu.DM.01G012790.1 | 191 | 21.29 | 9.37 | 24.95 | 77.12 | -0.019 | 1-24 | - | Cell wall. Chloroplast Mitochondrion |
| StDIR4 | Soltu.DM.01G019610.1 | 174 | 19.40 | 6.5 | 10.61 | 99.71 | 0.21 | - | - | Cell membrane |
| StDIR5 | Soltu.DM.01G050810.1 | 127 | 13.56 | 5.6 | 9.14 | 92.13 | 0.231 | 1-20 | - | Cell wall |
| StDIR6 | Soltu.DM.01G050820.1 | 188 | 20.76 | 9.1 | 31.15 | 90.27 | 0.062 | 1-22 | - | Cell membrane. Cell wall |
| StDIR7 | Soltu.DM.02G003630.1 | 190 | 21.10 | 8.76 | 27.85 | 92.37 | -0.015 | 1-22 | - | Cell membrane |
| StDIR8 | Soltu.DM.02G003640.1 | 185 | 20.57 | 9.1 | 25.39 | 91.19 | 0.001 | 1-17 | - | Cell membrane |
| StDIR9 | Soltu.DM.02G022680.1 | 318 | 34.60 | 9.56 | 27.65 | 91.01 | 0.041 | - | 289 315 | Endoplasmic reticulum |
| StDIR10 | Soltu.DM.04G003460.1 | 185 | 20.66 | 9.58 | 32.65 | 101.84 | 0.038 | - | 25 | Cell membrane |
| StDIR11 | Soltu.DM.04G007230.1 | 335 | 35.58 | 5.17 | 35.6 | 83.52 | -0.05 | 1-30 | - | Cell membrane |
| StDIR12 | Soltu.DM.05G023520.1 | 137 | 14.84 | 9.45 | 29.19 | 95.33 | 0.299 | 1-28 | - | Chloroplast. Peroxisome |
| StDIR13 | Soltu.DM.05G025340.1 | 400 | 41.21 | 4.29 | 41.77 | 88.55 | 0.023 | 1-30 | - | Cell membrane |
| StDIR14 | Soltu.DM.05G025350.1 | 240 | 26.46 | 6.34 | 34.48 | 78.38 | -0.238 | 1-25 | - | Nucleus |
| StDIR15 | Soltu.DM.06G015540.1 | 188 | 20.75 | 6.13 | 46.29 | 78.4 | 0.116 | 1-25 | - | Cell membrane |
| StDIR16 | Soltu.DM.06G031100.1 | 329 | 33.10 | 4.66 | 24.04 | 86.69 | 0.235 | 1-30 | - | Cell membrane |
| StDIR17 | Soltu.DM.07G013170.1 | 187 | 20.55 | 9.49 | 22.41 | 93.8 | 0.195 | 1-22 | - | Chloroplast |
| StDIR18 | Soltu.DM.07G017590.1 | 177 | 19.60 | 6.36 | 19.62 | 103.05 | 0.193 | 1-22 | 127 | Cell membrane Mitochondrion |
| StDIR19 | Soltu.DM.08G028530.1 | 189 | 21.53 | 6.35 | 25.21 | 70.69 | -0.086 | 1-24 | - | Cell membrane |
| StDIR20 | Soltu.DM.08G028540.1 | 189 | 21.16 | 6.9 | 20.34 | 91.9 | 0.1 | 1-26 | - | Cell membrane. Cell wall. Chloroplast |
| StDIR21 | Soltu.DM.09G028090.1 | 246 | 25.71 | 5.02 | 31.88 | 87.24 | 0.178 | 1-17 | 145 222 | Cell membrane |
| StDIR22 | Soltu.DM.10G004800.1 | 190 | 21.33 | 9.73 | 33.17 | 87.74 | 0.017 | 1-22 | 26 | Cell membrane |
| StDIR23 | Soltu.DM.10G017030.1 | 184 | 20.24 | 9.21 | 23.79 | 96.9 | 0.188 | 1-20 | 59 | Cell membrane. Cell wall. Chloroplast. Mitochondrion |
| StDIR24 | Soltu.DM.10G017040.1 | 194 | 20.93 | 9.27 | 20.18 | 101.6 | 0.176 | 1-26 | - | Cell membrane. Cell wall. Chloroplast |
| StDIR25 | Soltu.DM.10G017050.1 | 128 | 14.03 | 9.7 | 14.52 | 75.47 | 0.008 | 1-22 | 128 | Cell membrane |
| StDIR26 | Soltu.DM.10G017060.1 | 173 | 19.23 | 5.45 | 22.87 | 77.23 | -0.086 | - | - | Cell wall |
| StDIR27 | Soltu.DM.10G017070.1 | 194 | 20.90 | 9.3 | 17.68 | 105.62 | 0.213 | 1-26 | - | Cell membrane |
| StDIR28 | Soltu.DM.10G017090.1 | 193 | 21.18 | 9.52 | 16.09 | 82.28 | 0.069 | 1-23 | 75 | Cell membrane |
| StDIR29 | Soltu.DM.10G017100.1 | 174 | 19.35 | 5.81 | 20.61 | 79.02 | -0.113 | - | - | Cell wall |
| StDIR30 | Soltu.DM.10G017110.1 | 190 | 20.95 | 8.63 | 18.93 | 89.16 | 0.112 | 1-20 | 72 | Cell membrane. Cell wall |
| StDIR31 | Soltu.DM.10G017130.1 | 124 | 13.52 | 9.43 | 22.84 | 80.16 | 0.198 | - | - | Cell membrane |
| StDIR32 | Soltu.DM.10G017170.1 | 184 | 20.29 | 9.63 | 28.12 | 92.17 | 0.167 | 1-20 | - | Cell membrane. Cell wall |
| StDIR33 | Soltu.DM.11G026480.1 | 336 | 34.33 | 4.74 | 33.36 | 84.49 | 0.136 | 1-31 | 186 | Cell membrane |
| StDIR34 | Soltu.DM.12G003100.1 | 247 | 25.75 | 5.26 | 29.03 | 91.62 | 0.225 | 1-25 | 146 223 | Cell membrane |
表1 马铃薯DIR基因序列分析
Table 1 Sequence analysis of DIRs in potato
| 基因名Gene name | 基因ID Gene ID | 蛋白长度Length of protein/aa | 分子量Molecular weight/kD | 等电点pI | 不稳定系数Instability index | 脂肪系数Aliphatic index | 亲水性平均值Grand average of hydropathicity | 信号肽Signal peptide/aa | N-糖基化位点N-Glyc position | 亚细胞定位Subcellular location |
|---|---|---|---|---|---|---|---|---|---|---|
| StDIR1 | Soltu.DM.01G001610.1 | 192 | 21.26 | 9.76 | 31.2 | 100.47 | 0.167 | 1-24 | 41 | Chloroplast |
| StDIR2 | Soltu.DM.01G012660.1 | 189 | 21.17 | 9.57 | 28 | 86.19 | 0.155 | 1-24 | - | Cell membrane. Chloroplast |
| StDIR3 | Soltu.DM.01G012790.1 | 191 | 21.29 | 9.37 | 24.95 | 77.12 | -0.019 | 1-24 | - | Cell wall. Chloroplast Mitochondrion |
| StDIR4 | Soltu.DM.01G019610.1 | 174 | 19.40 | 6.5 | 10.61 | 99.71 | 0.21 | - | - | Cell membrane |
| StDIR5 | Soltu.DM.01G050810.1 | 127 | 13.56 | 5.6 | 9.14 | 92.13 | 0.231 | 1-20 | - | Cell wall |
| StDIR6 | Soltu.DM.01G050820.1 | 188 | 20.76 | 9.1 | 31.15 | 90.27 | 0.062 | 1-22 | - | Cell membrane. Cell wall |
| StDIR7 | Soltu.DM.02G003630.1 | 190 | 21.10 | 8.76 | 27.85 | 92.37 | -0.015 | 1-22 | - | Cell membrane |
| StDIR8 | Soltu.DM.02G003640.1 | 185 | 20.57 | 9.1 | 25.39 | 91.19 | 0.001 | 1-17 | - | Cell membrane |
| StDIR9 | Soltu.DM.02G022680.1 | 318 | 34.60 | 9.56 | 27.65 | 91.01 | 0.041 | - | 289 315 | Endoplasmic reticulum |
| StDIR10 | Soltu.DM.04G003460.1 | 185 | 20.66 | 9.58 | 32.65 | 101.84 | 0.038 | - | 25 | Cell membrane |
| StDIR11 | Soltu.DM.04G007230.1 | 335 | 35.58 | 5.17 | 35.6 | 83.52 | -0.05 | 1-30 | - | Cell membrane |
| StDIR12 | Soltu.DM.05G023520.1 | 137 | 14.84 | 9.45 | 29.19 | 95.33 | 0.299 | 1-28 | - | Chloroplast. Peroxisome |
| StDIR13 | Soltu.DM.05G025340.1 | 400 | 41.21 | 4.29 | 41.77 | 88.55 | 0.023 | 1-30 | - | Cell membrane |
| StDIR14 | Soltu.DM.05G025350.1 | 240 | 26.46 | 6.34 | 34.48 | 78.38 | -0.238 | 1-25 | - | Nucleus |
| StDIR15 | Soltu.DM.06G015540.1 | 188 | 20.75 | 6.13 | 46.29 | 78.4 | 0.116 | 1-25 | - | Cell membrane |
| StDIR16 | Soltu.DM.06G031100.1 | 329 | 33.10 | 4.66 | 24.04 | 86.69 | 0.235 | 1-30 | - | Cell membrane |
| StDIR17 | Soltu.DM.07G013170.1 | 187 | 20.55 | 9.49 | 22.41 | 93.8 | 0.195 | 1-22 | - | Chloroplast |
| StDIR18 | Soltu.DM.07G017590.1 | 177 | 19.60 | 6.36 | 19.62 | 103.05 | 0.193 | 1-22 | 127 | Cell membrane Mitochondrion |
| StDIR19 | Soltu.DM.08G028530.1 | 189 | 21.53 | 6.35 | 25.21 | 70.69 | -0.086 | 1-24 | - | Cell membrane |
| StDIR20 | Soltu.DM.08G028540.1 | 189 | 21.16 | 6.9 | 20.34 | 91.9 | 0.1 | 1-26 | - | Cell membrane. Cell wall. Chloroplast |
| StDIR21 | Soltu.DM.09G028090.1 | 246 | 25.71 | 5.02 | 31.88 | 87.24 | 0.178 | 1-17 | 145 222 | Cell membrane |
| StDIR22 | Soltu.DM.10G004800.1 | 190 | 21.33 | 9.73 | 33.17 | 87.74 | 0.017 | 1-22 | 26 | Cell membrane |
| StDIR23 | Soltu.DM.10G017030.1 | 184 | 20.24 | 9.21 | 23.79 | 96.9 | 0.188 | 1-20 | 59 | Cell membrane. Cell wall. Chloroplast. Mitochondrion |
| StDIR24 | Soltu.DM.10G017040.1 | 194 | 20.93 | 9.27 | 20.18 | 101.6 | 0.176 | 1-26 | - | Cell membrane. Cell wall. Chloroplast |
| StDIR25 | Soltu.DM.10G017050.1 | 128 | 14.03 | 9.7 | 14.52 | 75.47 | 0.008 | 1-22 | 128 | Cell membrane |
| StDIR26 | Soltu.DM.10G017060.1 | 173 | 19.23 | 5.45 | 22.87 | 77.23 | -0.086 | - | - | Cell wall |
| StDIR27 | Soltu.DM.10G017070.1 | 194 | 20.90 | 9.3 | 17.68 | 105.62 | 0.213 | 1-26 | - | Cell membrane |
| StDIR28 | Soltu.DM.10G017090.1 | 193 | 21.18 | 9.52 | 16.09 | 82.28 | 0.069 | 1-23 | 75 | Cell membrane |
| StDIR29 | Soltu.DM.10G017100.1 | 174 | 19.35 | 5.81 | 20.61 | 79.02 | -0.113 | - | - | Cell wall |
| StDIR30 | Soltu.DM.10G017110.1 | 190 | 20.95 | 8.63 | 18.93 | 89.16 | 0.112 | 1-20 | 72 | Cell membrane. Cell wall |
| StDIR31 | Soltu.DM.10G017130.1 | 124 | 13.52 | 9.43 | 22.84 | 80.16 | 0.198 | - | - | Cell membrane |
| StDIR32 | Soltu.DM.10G017170.1 | 184 | 20.29 | 9.63 | 28.12 | 92.17 | 0.167 | 1-20 | - | Cell membrane. Cell wall |
| StDIR33 | Soltu.DM.11G026480.1 | 336 | 34.33 | 4.74 | 33.36 | 84.49 | 0.136 | 1-31 | 186 | Cell membrane |
| StDIR34 | Soltu.DM.12G003100.1 | 247 | 25.75 | 5.26 | 29.03 | 91.62 | 0.225 | 1-25 | 146 223 | Cell membrane |
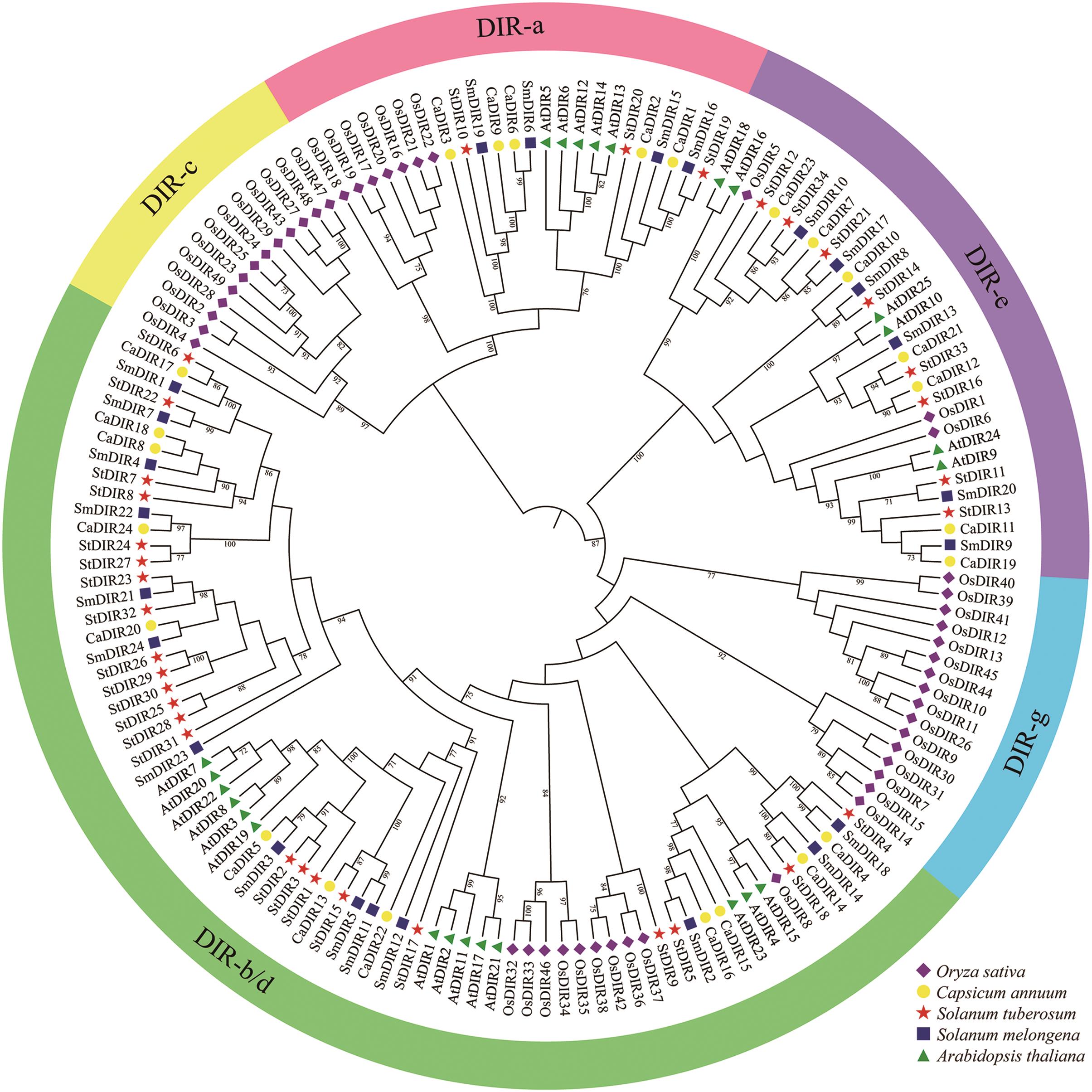
图2 马铃薯StDIRs与其它物种DIRs的系统发育关系不同DIR亚族用不同颜色表示。不同形状图标代表不同物种,菱形、圆形、五角星、正方形和三角形分别代表水稻(Oryza sativa)、辣椒(Capsicum annuum)、马铃薯(Solanum tuberosum)、茄子(Solanum melongena)和拟南芥(Arabidopsis thaliana)
Fig. 2 Phylogenetic relationship of StDIRs from potato and DIRs from other plant speciesDifferent groups of DIRs are represented by different colors. Different icons indicate different species, O. sativa (Os), C. annuum (Ca), S. tuberosum (St), S. melongena (Sm), and A. thaliana (At) is indicated by rhombus, circle, pentagram, square, and triangle
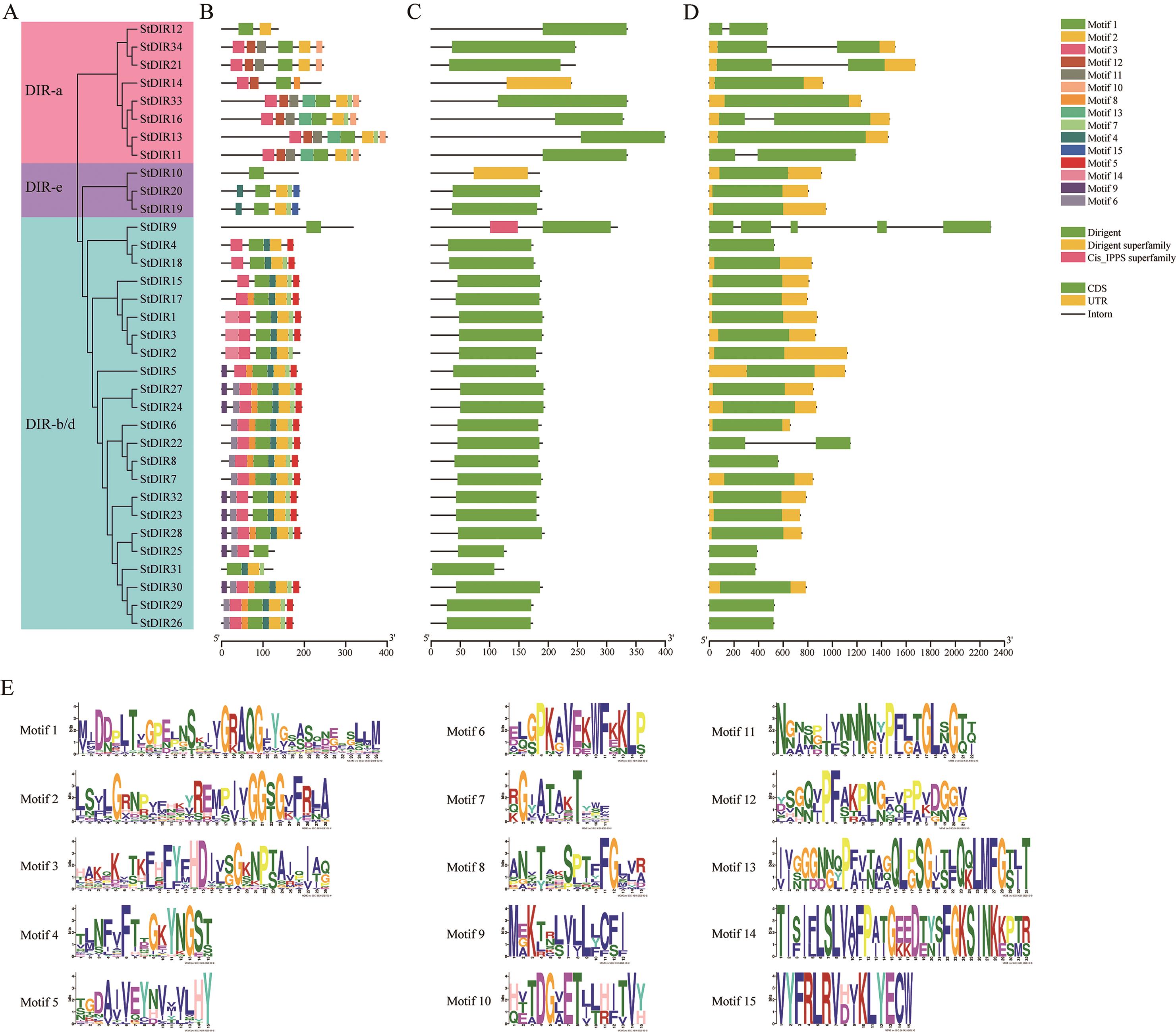
图3 马铃薯DIRs的保守基序、结构域和基因结构分析A:StDIRs系统发育树。B:StDIR蛋白保守基序分布。C:保守的DIR结构域位置。D:StDIR基因结构。E:每个motif的序列信息
Fig. 3 Analysis of conserved motifs, domains and gene structure of DIRs in potatoA: Phylogenetic tree of StDIRs. B: Distribution of conserved motifs of StDIR proteins. C: The location of the conserved DIR domain. D: Structure of StDIR gene. E: Sequence information for each motif

图4 马铃薯物种内DIR基因共线性分析蓝色弧线连接串联复制基因对,红色弧线连接片段复制基因对。圆环从内到外依次为染色体、未知碱基分布情况、GC含量的分布变化情况、全基因组基因密度统计
Fig. 4 Collinearity analysis of DIR genes in potatoThe blue refers to links tandem duplication gene pairs, red refers to links segmental duplication gene pairs. The chromosomes, distributions of unknown bases, distributions of GC content, and the genome-wide gene density are demonstrated by rings from the inside to the outside, respectively
| 1 | Davin LB, Wang HB, Crowell AL, et al. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center [J]. Science, 1997, 275(5298): 362-366. |
| 2 | Burlat V, Kwon M, Davin LB, et al. Dirigent proteins and dirigent sites in lignifying tissues [J]. Phytochemistry, 2001, 57(6): 883-897. |
| 3 | Ralph S, Park JY, Bohlmann J, et al. Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.) [J]. Plant Mol Biol, 2006, 60(1): 21-40. |
| 4 | Davin LB, Lewis NG. Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis [J]. Plant Physiol, 2000, 123(2): 453-462. |
| 5 | Ralph SG, Jancsik S, Bohlmann J. Dirigent proteins in conifer defense Ⅱ extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.) [J]. Phytochemistry, 2007, 68(14): 1975-1991. |
| 6 | Davin LB, Jourdes M, Patten AM, et al. Dissection of lignin macromolecular configuration and assembly: comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis [J]. Nat Prod Rep, 2008, 25(6):1015-1090. |
| 7 | Wu RH, Wang LL, Wang Z, et al. Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica [J]. Prog Nat Sci, 2009, 19(3): 347-352. |
| 8 | Li Q, Chen JF, Xiao Y, et al. The dirigent multigene family in Isatis indigotica: gene discovery and differential transcript abundance [J]. BMC Genomics, 2014, 15(1): 388. |
| 9 | Halls SC, Davin LB, Kramer DM, et al. Kinetic study of coniferyl alcohol radical binding to the (+)-pinoresinol forming dirigent protein [J]. Biochemistry, 2004, 43(9): 2587-2595. |
| 10 | Pellegrini N, Valtueña S, Ardigò D, et al. Intake of the plant lignans matairesinol, secoisolariciresinol, pinoresinol, and lariciresinol in relation to vascular inflammation and endothelial dysfunction in middle age-elderly men and post-menopausal women living in Northern Italy [J]. Nutr Metab Cardiovasc Dis, 2010, 20(1): 64-71. |
| 11 | Ascacio-Valdés J, Buenrostro-Figueroa JJ, Aguilera-Carbó A, et al. Ellagitannins: Biosynthesis, biodegradation and biological properties [J]. J Med Plants Res, 2011, 5: 4696-4703. |
| 12 | Zhan ZJ, Ying YM, Ma LF, et al. Natural disesquiterpenoids [J]. Nat Prod Rep, 2011, 28(3): 594-629. |
| 13 | Rivière C, Pawlus AD, Mérillon JM. Natural stilbenoids: distribution in the plant Kingdom and chemotaxonomic interest in Vitaceae [J]. Nat Prod Rep, 2012, 29(11): 1317-1333. |
| 14 | Satake H, Koyama T, Bahabadi SE, et al. Essences in metabolic engineering of lignan biosynthesis [J]. Metabolites, 2015, 5(2): 270-290. |
| 15 | Paniagua C, Bilkova A, Jackson P, et al. Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure [J]. J Exp Bot, 2017, 68(13): 3287-3301. |
| 16 | Umezawa T, Davin LB, Yamamoto E, et al. Lignan biosynthesis in Forsythia species [J]. J Chem Soc, Chem Commun, 1990(20): 1405. |
| 17 | Cheng X, Su XQ, Muhammad A, et al. Molecular characterization, evolution, and expression profiling of the Dirigent (DIR) family genes in Chinese white pear (Pyrus bretschneideri) [J]. Front Genet, 2018, 9: 136. |
| 18 | Khan A, Li RJ, Sun JT, et al. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses [J]. Sci Rep, 2018, 8(1): 5500. |
| 19 | Liu ZW, Wang XF, Sun ZW, et al. Evolution, expression and functional analysis of cultivated allotetraploid cotton DIR genes [J]. BMC Plant Biol, 2021, 21(1): 89. |
| 20 | Hosmani PS, Kamiya T, Danku J, et al. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root [J]. Proc Natl Acad Sci USA, 2013, 110(35): 14498-14503. |
| 21 | Corbin C, Drouet S, Markulin L, et al. A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: from gene identification and evolution to differential regulation [J]. Plant Mol Biol, 2018, 97(1/2): 73-101. |
| 22 | Chou KC, Shen HB. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization [J]. PLoS One, 2010, 5(6): e11335. |
| 23 | Gupta R, Jung E, Brunak S. Prediction of N-glycosylation sites in human proteins [J]. 2004, 46: 203-206. |
| 24 | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| 25 | Zhang KJ, Xing WJ, Sheng SA, et al. Genome-wide identification and expression analysis of eggplant DIR gene family in response to biotic and abiotic stresses [J]. Horticulturae, 2022, 8(8): 732. |
| 26 | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching [J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| 27 | Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server [J]. Bioinformatics, 2015, 31(8): 1296-1297. |
| 28 | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| 29 | Thamil Arasan SK, Park JI, Ahmed NU, et al. Characterization and expression analysis of dirigent family genes related to stresses in Brassica [J]. Plant Physiol Biochem, 2013, 67: 144-153. |
| 30 | Liao YR, Liu SB, Jiang YY, et al. Genome-wide analysis and environmental response profiling of dirigent family genes in rice (Oryza sativa) [J]. Genes Genom, 2017, 39(1): 47-62. |
| 31 | Song M, Peng XY. Genome-wide identification and characterization of DIR genes in Medicago truncatula [J]. Biochem Genet, 2019, 57(4): 487-506. |
| 32 | Li LL, Sun WB, Zhou PJ, et al. Genome-wide characterization of dirigent proteins in Populus: gene expression variation and expression pattern in response to Marssonina Brunnea and phytohormones [J]. Forests, 2021, 12(4): 507. |
| 33 | Ma XF, Xu WY, Liu T, et al. Functional characterization of soybean (Glycine max) DIRIGENT genes reveals an important role of GmDIR27 in the regulation of pod dehiscence [J]. Genomics, 2021, 113(1 Pt 2): 979-990. |
| 34 | Yadav V, Wang ZY, Yang XZ, et al. Comparative analysis, characterization and evolutionary study of dirigent gene family in Cucurbitaceae and expression of novel dirigent peptide against powdery mildew stress [J]. Genes, 2021, 12(3): 326. |
| 35 | 张洪伟. 马铃薯晚疫病抗性相关基因StDIR1和StDIR2的分离和表达 [D]. 保定: 河北大学, 2012. |
| Zhang HW. Isolation and expression of StDIR1 and StDIR2 genes related to potato late blight resistance [D]. Baoding: Hebei University, 2012. | |
| 36 | Liu JG, Stipanovic RD, Bell AA, et al. Stereoselective coupling of hemigossypol to form (+)-gossypol in moco cotton is mediated by a dirigent protein [J]. Phytochemistry, 2008, 69(18): 3038-3042. |
| 37 | Effenberger I, Zhang B, Li L, et al. Dirigent proteins from cotton (Gossypium sp.) for the atropselective synthesis of gossypol [J]. Angew Chem Int Ed, 2015, 54(49): 14660-14663. |
| 38 | Sahani P, Ujinwal M, Singh S, et al. Genome wide in silico characterization of dirigent protein family in flax (Linum usitatissmum l.) [J]. Plant Archives, 2018, 18: 61-68. |
| 39 | Ma RF, Huang B, Chen JL, et al. Genome-wide identification and expression analysis of dirigent-jacalin genes from plant chimeric lectins in Moso bamboo (Phyllostachys edulis) [J]. PLoS One, 2021, 16(3): e0248318. |
| 40 | Kim MK, Jeon JH, Fujita M, et al. The western red cedar (Thuja plicata) 8-8' DIRIGENT family displays diverse expression patterns and conserved monolignol coupling specificity [J]. Plant Mol Biol, 2002, 49(2): 199-214. |
| 41 | 马金洋, 杨瑾冬, 李卿, 等. 丹参Dirigent基因家族的发现与生物信息学分析 [J]. 基因组学与应用生物学, 2017, 36(4): 1594-1610. |
| Ma JY, Yang JD, Li Q, et al. Discovery and bioinformatics analysis of dirigent multigene family in Salvia miltiorrhiza [J]. Genom Appl Biol, 2017, 36(4): 1594-1610. | |
| 42 | 刘正文. 棉花纤维发育相关基因挖掘及DIR和GH9基因家族研究 [D]. 保定: 河北农业大学, 2021. |
| Liu ZW. Mining genes related to cotton fiber development and study on DIR and GH9 gene family [D]. Baoding: Hebei Agricultural University, 2021. | |
| 43 | 穰中文, 周清明. 水稻dirigent基因家族生物信息学分析 [J]. 湖南农业大学学报: 自然科学版, 2013, 39(2): 111-120. |
| Rang ZW, Zhou QM. Bioinformatic analysis of the dirigent gene family in rice [J]. J Hunan Agric Univ Nat Sci, 2013, 39(2): 111-120. | |
| 44 | Zhu LF, Zhang XL, Tu LL, et al. Isolation and characterization of two novel dirigent-like genes highly induced in cotton (Gossypium barbadense and G. hirsutum) after infection by Verticillium dahliae [J]. J Plant Pathol, 2007, 89: 41-45. |
| 45 | 关瑞攀. Dirigent基因参与三七-茄腐镰刀菌互作的分子机理研究 [D]. 昆明: 昆明理工大学, 2018. |
| Guan RP. Molecular mechanism of Dirigent genes involved in the interaction of Panax notoginseng-Fusarium solani [D]. Kunming: Kunming University of Science and Technology, 2018. | |
| 46 | Guo JL, Xu LP, Fang JP, et al. A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses [J]. Plant Cell Rep, 2012, 31(10): 1801-1812. |
| 47 | Shi YQ, Shen YR, Ahmad B, et al. Genome-wide identification and expression analysis of dirigent gene family in strawberry (Fragaria vesca) and functional characterization of FvDIR13 [J]. Sci Hortic, 2022, 297: 110913. |
| [1] | 颜伟, 陈慧婷, 叶青, 刘广超, 刘新, 侯丽霞. 葡萄HCT基因家族鉴定及其对低温胁迫的响应[J]. 生物技术通报, 2025, 41(2): 175-186. |
| [2] | 匡健华, 程志鹏, 赵永晶, 杨洁, 陈润乔, 陈龙清, 胡慧贞. 激素和非生物胁迫下荷花GH3基因家族的表达分析[J]. 生物技术通报, 2025, 41(2): 221-233. |
| [3] | 黄颖, 遇文婧, 刘雪峰, 刁桂萍. 山新杨谷胱甘肽转移酶基因的生物信息学与表达模式分析[J]. 生物技术通报, 2025, 41(2): 248-256. |
| [4] | 杨涌, 袁国梅, 康肖肖, 刘亚明, 王东升, 张海娥. 板栗SWEET基因家族成员的鉴定及表达分析[J]. 生物技术通报, 2025, 41(2): 257-269. |
| [5] | 杨涌, 曹蕊, 康肖肖, 刘静, 王旋, 张海娥. 板栗类黄酮合成通路13个基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(2): 270-283. |
| [6] | 李明, 刘祥宇, 王益娜, 和四梅, 沙本才. 紫金龙异紫堇定生物合成相关6-OMT基因克隆与功能表征[J]. 生物技术通报, 2025, 41(2): 309-320. |
| [7] | 葛仕杰, 刘怡德, 张华东, 宁强, 朱展望, 王书平, 刘易科. 小麦蛋白质二硫键异构酶基因家族的鉴定与表达[J]. 生物技术通报, 2025, 41(2): 85-96. |
| [8] | 殷缘, 程爽, 刘定豪, 邓晓霞, 李凯月, 王竞红, 蔺吉祥. 外源过氧化氢(H2O2)影响非生物胁迫下植物生长与生理代谢机制的研究进展[J]. 生物技术通报, 2025, 41(1): 1-13. |
| [9] | 杜品廷, 吴国江, 王振国, 李岩, 周伟, 周亚星. 高粱CPP基因家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 132-142. |
| [10] | 武志健, 刘广洋, 林志豪, 盛彬, 陈鸽, 许晓敏, 王军伟, 徐东辉. 蔬菜种子萌发的纳米调控及其机制研究进展[J]. 生物技术通报, 2025, 41(1): 14-24. |
| [11] | 李禹欣, 李苗, 杜晓芬, 韩康妮, 连世超, 王军. 谷子SiSAP基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(1): 143-156. |
| [12] | 王子傲, 田瑞, 崔永梅, 白羿雄, 姚晓华, 安立昆, 吴昆仑. 青稞HvnJAZ4的生物信息学和表达模式分析[J]. 生物技术通报, 2025, 41(1): 173-185. |
| [13] | 李彩霞, 李艺, 穆宏秀, 林俊轩, 白龙强, 孙美华, 苗妍秀. 中国南瓜bHLH转录因子家族的鉴定与生物信息学分析[J]. 生物技术通报, 2025, 41(1): 186-197. |
| [14] | 孔青洋, 张晓龙, 李娜, 张晨洁, 张雪云, 于超, 张启翔, 罗乐. 单叶蔷薇GRAS转录因子家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 210-220. |
| [15] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||