








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 139-149.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0557
倪莹1( ), 李雷1, 汪进萱1, 马波1, 孟昕2, 冷平生1,3, 吴静1,3(
), 李雷1, 汪进萱1, 马波1, 孟昕2, 冷平生1,3, 吴静1,3( ), 胡增辉1,3(
), 胡增辉1,3( )
)
收稿日期:2025-05-31
出版日期:2026-01-26
发布日期:2026-02-04
通讯作者:
吴静,女,博士,副教授,研究方向 :园林植物与观赏园艺;E-mail: wjmxy1988@126.com作者简介:倪莹,女,硕士研究生,研究方向 :园林植物与观赏园艺;E-mail: 18810602920@163.com
基金资助:
NI Ying1( ), LI Lei1, WANG Jin-xuan1, MA Bo1, MENG Xin2, LENG Ping-sheng1,3, WU Jing1,3(
), LI Lei1, WANG Jin-xuan1, MA Bo1, MENG Xin2, LENG Ping-sheng1,3, WU Jing1,3( ), HU Zeng-hui1,3(
), HU Zeng-hui1,3( )
)
Received:2025-05-31
Published:2026-01-26
Online:2026-02-04
摘要:
目的 4-香豆酸∶辅酶A连接酶(4-coumarate: CoA lig-ase, 4CL)是苯丙烷代谢途径中的核心酶,克隆并探究4CL基因在紫丁香(Syringa oblata)花青苷合成中的功能。 方法 基于紫丁香基因组及转录组数据筛选并克隆出So4CL基因,对其编码的蛋白进行生物信息学分析和亚细胞定位;利用RT-qPCR研究So4CL在不同花期及不同组织的相对表达量;构建So4CL过表达和沉默载体,利用农杆菌侵染法分别转化紫丁香和烟草(Nicotiana benthamiana),观察其表型变化并测定花青苷含量;通过RT-qPCR检测花青苷生物合成途径中So4CL上下游基因的表达水平。 结果 So4CL基因CDS区全长为1 659 bp,编码552个氨基酸,具有BOX I和BOX Ⅱ保守结构域。系统进化分析显示,紫丁香So4CL与水曲柳(Fraxinus mandshurica)的4CL蛋白相似性最高,为97%。RT-qPCR分析结果显示,随着花发育,紫丁香花瓣褪色,So4CL的表达整体呈降低的趋势;So4CL在盛花期中的根、茎、叶、花中均有表达。亚细胞定位分析显示,So4CL主要存在于细胞质上。过表达So4CL的紫丁香花瓣着色明显,花青苷含量显著升高,烟草叶片出现砖红色变化;沉默So4CL后花瓣显著褪色,花青苷含量显著降低;RT-qPCR分析表明,瞬时转化So4CL会影响花青苷生物合成途径中SoPAL、SoCHS、SoDFR和SoUFGT的表达。 结论 So4CL在紫丁香花瓣花青苷合成中发挥着重要作用,是合成关键基因。
倪莹, 李雷, 汪进萱, 马波, 孟昕, 冷平生, 吴静, 胡增辉. 紫丁香So4CL的克隆及功能分析[J]. 生物技术通报, 2026, 42(1): 139-149.
NI Ying, LI Lei, WANG Jin-xuan, MA Bo, MENG Xin, LENG Ping-sheng, WU Jing, HU Zeng-hui. Cloning and Functional Analysis of So4CL Gene in Syringa oblata[J]. Biotechnology Bulletin, 2026, 42(1): 139-149.
引物用途 Primer function | 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) |
|---|---|---|
克隆基因 Gene cloning | So4CL-F So4CL-R | ATGGAGACTAAGACGACGCAAGAAGA TCAATTGGGAACACCTGCAGCTAATC |
实时荧光定量PCR RT-qPCR | qSoActin-F qSoActin-R qSo4CL-F qSo4CL-R qSoPAL-F qSoPAL-R qSoCHS-F qSoCHS-R qSoDFR-F qSoDFR-R qSoUFGT-F qSoUFGT-R | TGGAATGTGCTGAGAGATGC TGCTGACCGTATGAGCAAAG CTTGCGGTCTGTCTTGTCTG TTCCGTCAAACCATAGCCCT TGGACTATGGCTTCAAGGGGR CTCGCCGAAATCAAACCCAA TCTCGTAGTGTGCTCGGAAA GTCAGAACCCACAATCACGG GCATTGGAAGCGGCTAAAGA CCAGTGATAGGCGAAAGTGC TACCACCAGAATAGAGT AGAAGTATTGCCAGAAG |
基因过表达 Gene overexpression | PRI101-So4CL-F PRI101-So4CL-R | ATGCCCGTCGACCCCGGGGGCGGAACTCAACCAGC CTCACCATGGATCCGGTACCTAGTTTAGATACGGCAAGCTTTATTAGATCCTTCC |
基因沉默 Gene silencing | TRV2-So4CL-F TRV2-So4CL-R | GTCCAGTCCTGGCCTCGTCGGCCATGGCGGAACTCAACCAGC GACCACAAGTGGCCAGACTGGCCAAGCTGAGCTGATGCCCG |
So4CL-SubN-F So4CL-SubN-R | AGTGGTCTCTGTCCAGTCCTATGGAGACTAAGACGACGCAAGAAGA GGTCTCAGCAGACCACAAGTATTGGGAACACCTGCAGCTAATC |
表1 引物序列
Table 1 Primer sequences
引物用途 Primer function | 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) |
|---|---|---|
克隆基因 Gene cloning | So4CL-F So4CL-R | ATGGAGACTAAGACGACGCAAGAAGA TCAATTGGGAACACCTGCAGCTAATC |
实时荧光定量PCR RT-qPCR | qSoActin-F qSoActin-R qSo4CL-F qSo4CL-R qSoPAL-F qSoPAL-R qSoCHS-F qSoCHS-R qSoDFR-F qSoDFR-R qSoUFGT-F qSoUFGT-R | TGGAATGTGCTGAGAGATGC TGCTGACCGTATGAGCAAAG CTTGCGGTCTGTCTTGTCTG TTCCGTCAAACCATAGCCCT TGGACTATGGCTTCAAGGGGR CTCGCCGAAATCAAACCCAA TCTCGTAGTGTGCTCGGAAA GTCAGAACCCACAATCACGG GCATTGGAAGCGGCTAAAGA CCAGTGATAGGCGAAAGTGC TACCACCAGAATAGAGT AGAAGTATTGCCAGAAG |
基因过表达 Gene overexpression | PRI101-So4CL-F PRI101-So4CL-R | ATGCCCGTCGACCCCGGGGGCGGAACTCAACCAGC CTCACCATGGATCCGGTACCTAGTTTAGATACGGCAAGCTTTATTAGATCCTTCC |
基因沉默 Gene silencing | TRV2-So4CL-F TRV2-So4CL-R | GTCCAGTCCTGGCCTCGTCGGCCATGGCGGAACTCAACCAGC GACCACAAGTGGCCAGACTGGCCAAGCTGAGCTGATGCCCG |
So4CL-SubN-F So4CL-SubN-R | AGTGGTCTCTGTCCAGTCCTATGGAGACTAAGACGACGCAAGAAGA GGTCTCAGCAGACCACAAGTATTGGGAACACCTGCAGCTAATC |
蛋白名称 Protein name | 分子式 Formula | 相对分子质量 Molecular weight (Da) | 总原子数 Total number of atoms | 酸碱性氨基酸 Acid-base amino acid | 理论等电点 Theoretical pI | 脂溶指数 Aliphatic index | |
|---|---|---|---|---|---|---|---|
| Asp + Glu | Arg + Lys | ||||||
| So4CL | C2716H4383N709O804S18 | 60 411.03 | 8 630 | 57 | 61 | 8.54 | 102.28 |
表2 So4CL蛋白质理化性质分析
Table 2 Analysis of physical and chemical properties of So4CL protein
蛋白名称 Protein name | 分子式 Formula | 相对分子质量 Molecular weight (Da) | 总原子数 Total number of atoms | 酸碱性氨基酸 Acid-base amino acid | 理论等电点 Theoretical pI | 脂溶指数 Aliphatic index | |
|---|---|---|---|---|---|---|---|
| Asp + Glu | Arg + Lys | ||||||
| So4CL | C2716H4383N709O804S18 | 60 411.03 | 8 630 | 57 | 61 | 8.54 | 102.28 |

图3 So4CL与其他物种4CL蛋白序列比对分析黑色、红色、蓝色部分分别表示同源性=100%、≥75%、≥50%;红色框为SSGTTGLPKGV保守基序,绿色框为GEXXIXG保守基序
Fig. 3 Alignment and analysis of So4CL with 4CL protein sequences of other speciesThe black, red, and blue parts indicate homology =100%, ≥75%, ≥50% respectively. Red boxes are SSGTTGLPKGV conserved motifs, and green boxes are GEXXIXG conserved motifs
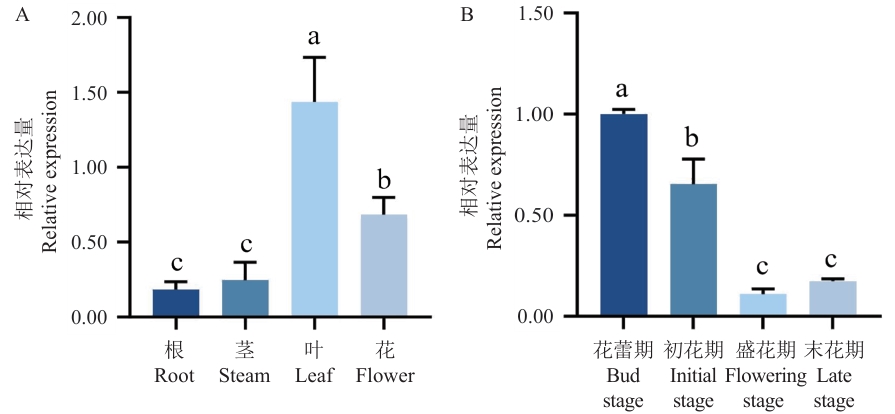
图5 So4CL在不同器官(A)和不同花发育阶段(B)的表达模式不同小写字母表示在P<0.05水平上差异显著
Fig. 5 Expression patterns of So4CL at different organs (A) and in different flower developmental stages (B)Different lowercase letters indicate significant differences at the P<0.05 level
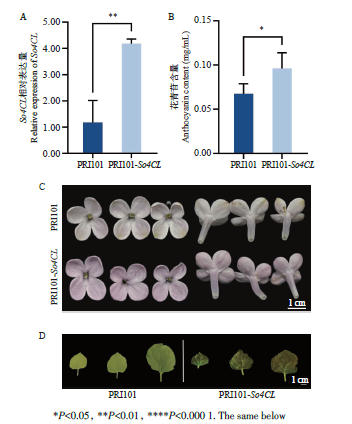
图7 紫丁香过表达So4CL植株的基因表达水平(A)、花青苷含量(B)、花色(C)和烟草过表达So4CL叶色变化(D)*P<0.05, **P<0.01, ****P<0.000 1. The same below
Fig. 7 Expressions of So4CL (A), anthocyanin contents (B), and petal color (C) in So4CL-overexpressed S. oblata and leaf color in So4CL-overexpressing N. benthamian (D)
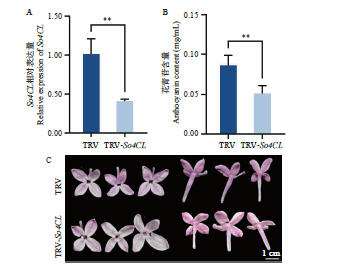
图8 紫丁香沉默So4CL 植株的基因表达水平(A)、花青苷含量(B)和花色(C)的变化
Fig. 8 Expressions of So4CL (A), anthocyanin contents(B), and petal color (C) in So4CL-silenced S. oblata
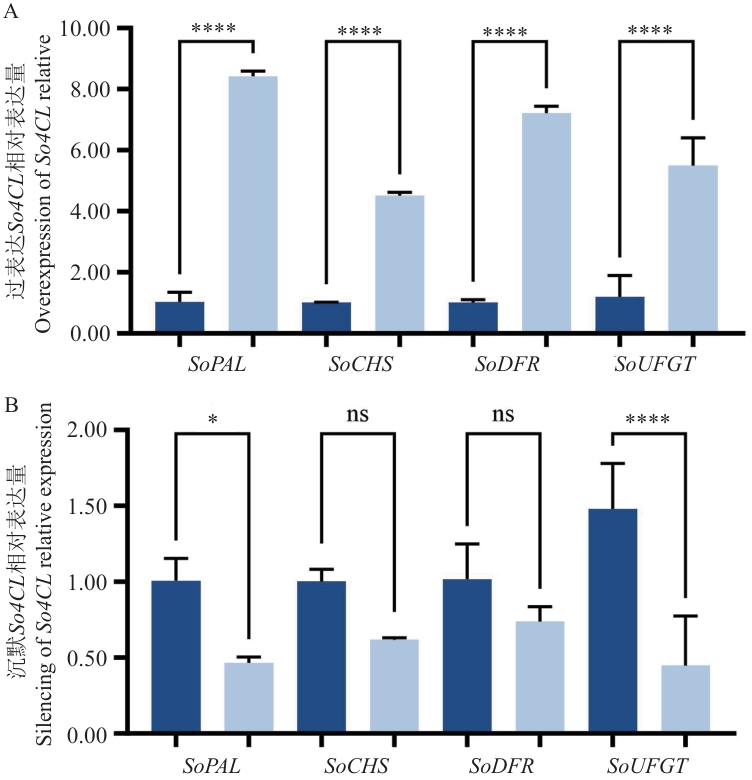
图9 瞬时过表达(A)和沉默(B)So4CL对上下游基因表达的影响
Fig. 9 Effects of transient overexpression (A) and silencing (B) of So4CL on the expressions of upstream and downstream genes
| [1] | Ma B, Wu J, Shi TL, et al. Lilac (Syringa oblata) genome provides insights into its evolution and molecular mechanism of petal color change [J]. Commun Biol, 2022, 5(1): 686. |
| [2] | 宋毓晔, 王渝, 朱千林, 等. 植物中花色苷来源及提取方法研究进展 [J]. 食品研究与开发, 2022, 43(16): 199-208. |
| Song YY, Wang Y, Zhu QL, et al. Research progress on sources and extraction methods of plant-based anthocyanin [J]. Food Res Dev, 2022, 43(16): 199-208. | |
| [3] | Zhu J, Wang YZ, Wang QY, et al. The combination of DNA methylation and positive regulation of anthocyanin biosynthesis by MYB and bHLH transcription factors contributes to the petal blotch formation in Xibei tree peony [J]. Hortic Res, 2023, 10(6): uhad100. |
| [4] | Deng CY, Wang JY, Lu CF, et al. CcMYB6-1 and CcbHLH1, two novel transcription factors synergistically involved in regulating anthocyanin biosynthesis in cornflower [J]. Plant Physiol Biochem, 2020, 151: 271-283. |
| [5] | 黄玲, 胡先梅, 梁泽慧, 等. 郁金香花青素合成酶基因TgANS的克隆与功能鉴定 [J]. 园艺学报, 2022, 49(9): 1935-1944. |
| Huang L, Hu XM, Liang ZH, et al. Cloning and function identification of anthocyanidin synthase gene TgANS in Tulipa gesneriana [J]. Acta Hortic Sin, 2022, 49(9): 1935-1944. | |
| [6] | 刘玥, 李月庆, 孟祥宇, 等. 大花君子兰查尔酮合酶基因CmCHS的克隆及其功能验证 [J]. 园艺学报, 2021, 48(10): 1847-1858. |
| Liu Y, Li YQ, Meng XY, et al. Cloning and functional characterization of chalcone synthase genes (CmCHS) from Clivia miniata [J]. Acta Hortic Sin, 2021, 48(10): 1847-1858. | |
| [7] | Osorio-Guarín JA, Gopaulchan D, Quanckenbush C, et al. Comparative transcriptomic analysis reveals key components controlling spathe color in Anthurium andraeanum (Hort.) [J]. PLoS One, 2021, 16(12): e0261364. |
| [8] | Rausher MD, Miller RE, Tiffin P. Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway [J]. Mol Biol Evol, 1999, 16(2): 266-274. |
| [9] | Chen XH, Wang HT, Li XY, et al. Molecular cloning and functional analysis of 4-Coumarate: CoA ligase 4 (4CL-like 1) from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis [J]. BMC Plant Biol, 2019, 19(1): 231. |
| [10] | Abdullah-Zawawi MR, Ahmad-Nizammuddin NF, Govender N, et al. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice [J]. Sci Rep, 2021, 11(1): 19678. |
| [11] | 申晚霞, 王志彬, 薛杨, 等. 柑橘4CL基因家族的结构及其功能分析 [J]. 园艺学报, 2019, 46(6): 1068-1078. |
| Shen WX, Wang ZB, Xue Y, et al. Characterization of 4-coumarate: CoA ligase (4CL) gene family in Citrus [J]. Acta Hortic Sin, 2019, 46(6): 1068-1078. | |
| [12] | Ma JY, Zuo DJ, Zhang XD, et al. Genome-wide identification analysis of the 4-Coumarate: CoA ligase (4CL) gene family expression profiles in Juglans regia and its wild relatives J. Mandshurica resistance and salt stress [J]. BMC Plant Biol, 2024, 24(1): 211. |
| [13] | 陈心仪, 吴成英, 贺海皓, 等. 滇水金凤4CL基因的克隆及表达分析 [J]. 福建农业学报, 2024, 39(1): 40-48. |
| Chen XY, Wu CY, He HH, et al. Cloning and expression of 4CLs in Impatiens uliginosa [J]. Fujian J Agric Sci, 2024, 39(1): 40-48. | |
| [14] | Li Y, Kim JI, Pysh L, et al. Four isoforms of Arabidopsis 4-coumarate: CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism [J]. Plant Physiol, 2015, 169(4): 2409-2421. |
| [15] | 张蕾, 林晓, 罗赟, 等. RNAi沉默Fa4CL基因对草莓果实花色苷代谢的影响 [J]. 果树学报, 2015, 32(3): 434-439, 522. |
| Zhang L, Lin X, Luo Y, et al. Influences of RNAi-induced Fa4CL silencing on anthocyanin metabolism in strawberry fruit [J]. J Fruit Sci, 2015, 32(3): 434-439, 522. | |
| [16] | Zhou XH, Jacobs TB, Xue LJ, et al. Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate: CoA ligase specificity and redundancy [J]. New Phytol, 2015, 208(2): 298-301. |
| [17] | 陈锐, 马道铖, 徐睆, 等. 野蔷薇4CL基因的克隆及表达分析 [J]. 北方园艺, 2022(17): 69-78. |
| Chen R, Ma DC, Xu H, et al. Cloning, sequencing and expression analysis of Rosa multiflora Rm4CL gene [J]. North Hortic, 2022(17): 69-78. | |
| [18] | 陈蒙, 刘海峰. 山葡萄4CL基因克隆与表达分析 [J]. 扬州大学学报: 农业与生命科学版, 2019, 40(5): 20-25. |
| Chen M, Liu HF. Expression of the 4CL gene isolated from Vitis amurensis in E. coli [J]. J Yangzhou Univ Agric Life Sci Ed, 2019, 40(5): 20-25. | |
| [19] | Chen XH, Su WL, Zhang H, et al. Fraxinus mandshurica 4-coumarate-CoA ligase 2 enhances drought and osmotic stress tolerance of tobacco by increasing coniferyl alcohol content [J]. Plant Physiol Biochem, 2020, 155: 697-708. |
| [20] | 徐靖, 林延慧, 王效宁, 等. 甘薯4-香豆酸辅酶A连接酶基因的生物信息学鉴定和表达分析 [J]. 西北植物学报, 2020, 40(4): 581-587. |
| Xu J, Lin YH, Wang XN, et al. Bioinformatic identification and expression analysis of 4CL genes in Ipomoea batatas [J]. Acta Bot Boreali Occidentalia Sin, 2020, 40(4): 581-587. | |
| [21] | 韩俊杰, 王萍, 孙淑荣, 等. 气候变化对黑龙江紫丁香物候期的影响 [J]. 中国农学通报, 2023, 39(26): 130-136. |
| Han JJ, Wang P, Sun SR, et al. Effects of climate change on phenology of lilac in Heilongjiang province [J]. Chin Agric Sci Bull, 2023, 39(26): 130-136. | |
| [22] | 范小峰, 刘秀丽, 卢晶. 紫丁香的组织培养和快速繁殖 [J]. 林业科技通讯, 2024(5): 51-54. |
| Fan XF, Liu XL, Lu J. Tissue culture and rapid propagation of Syringa oblata [J]. For Sci Technol, 2024(5): 51-54. | |
| [23] | 孙全航, 康璠, 何淼, 等. 镉胁迫对紫丁香种子萌发及幼苗生长的影响 [J]. 种子, 2024, 43(2): 88-94. |
| Sun QH, Kang F, He M, et al. Effects of cadmium stress on seed germination and seedling growth of Syringa oblata L [J]. Seed, 2024, 43(2): 88-94. | |
| [24] | 王蕊, 王羽, 李彦慧, 等. 华北紫丁香肉桂酸4-羟化酶(SoC4H)基因的克隆及表达分析 [J]. 分子植物育种, 2016, 14(8): 2025-2030. |
| Wang R, Wang Y, Li YH, et al. Cloning and expression analysis of cinnamic acid 4-hydroxylase gene from Syringa oblata lindl [J]. Mol Plant Breed, 2016, 14(8): 2025-2030. | |
| [25] | 王蕊, 李彦慧, 郑健. 紫丁香查尔酮异构酶基因SoCHI的克隆及表达分析 [J]. 植物资源与环境学报, 2018, 27(3): 11-17. |
| Wang R, Li YH, Zheng J. Cloning and expression analysis on Chalcone isomerase gene SoCHI in Syringa oblata [J]. J Plant Resour Environ, 2018, 27(3): 11-17. | |
| [26] | 李文芳. 苹果4CL3基因启动子的克隆及其在花青素合成中的功能分析[D]. 兰州: 甘肃农业大学, 2023. |
| Li YF. Cloning of 4CL3 gene promoter and functional analysis of its role in anthocyanin synthesis in Malus domestica [D]. Lanzhou: Gansu Agricultural University, 2023. | |
| [27] | 马馨馨, 许洋, 赵欢欢, 等. 番茄4CL基因家族鉴定和氮素处理下的表达分析 [J]. 生物技术通报, 2022, 38(4): 163-173. |
| Ma XX, Xu Y, Zhao HH, et al. Identification of tomato 4CL gene family and expression analysis under nitrogen treatment [J]. Biotechnol Bull, 2022, 38(4): 163-173. | |
| [28] | Wang Y, Dou Y, Wang R, et al. Molecular characterization and functional analysis of chalcone synthase from Syringa oblata Lindl. in the flavonoid biosynthetic pathway [J]. Gene, 2017, 635: 16-23. |
| [29] | 马波, 李雷, 胡增辉, 等. 紫丁香SoF3′H对花瓣中花青苷合成的功能解析 [J]. 园艺学报, 2024, 51(8): 1833-1843. |
| Ma B, Li L, Hu ZH, et al. Functional analysis of SoF3′H in the synthesis of anthocyanidin in Syringa oblata petals [J]. Acta Hortic Sin, 2024, 51(8): 1833-1843. | |
| [30] | Uhlmann A, Ebel J. Molecular cloning and expression of 4-coumarate: coenzyme A ligase, an enzyme involved in the resistance response of soybean (Glycine max L.) against pathogen attack [J]. Plant Physiol, 1993, 102(4): 1147-1156. |
| [31] | 钟秀来, 赵倩, 朱顺华, 等. 芹菜Ag4CL1基因克隆及功能分析 [J/OL]. 分子植物育种, 2023. . |
| Zhong XL, Zhao Q, Zhu SH, et al. Cloning and expression analysis of Ag4CL1 gene in Apium graveolens [J/OL]. Mol Plant Breed, 2023. . | |
| [32] | 余珠, 欧阳乐军, 赵永国, 等. 4-香豆酸辅酶A连接酶(4CL)的研究进展 [J/OL]. 分子植物育种, 2023. . |
| Yu Z, Ouyang LJ, Zhao YG, et al. Research progress of 4-coumaric acid coenzyme aligase (4CL) [J/OL]. Mol Plant Breed, 2023. . | |
| [33] | Cao Y, Hu SL, Huang SX, et al. Molecular cloning, expression pattern, and putative cis-acting elements of a 4-coumarate: CoA ligase gene in bamboo (Neosinocalamus affinis) [J]. Electron J Biotechnol, 2012, 15(5): 9. |
| [34] | Allina SM, Pri-Hadash A, Theilmann DA, et al. 4-Coumarate: coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes [J]. Plant Physiol, 1998, 116(2): 743-754. |
| [35] | Yan C, Li CL, Jiang MC, et al. Systematic characterization of gene families and functional analysis of PvRAS3 and PvRAS4 involved in rosmarinic acid biosynthesis in Prunella vulgaris [J]. Front Plant Sci, 2024, 15: 1374912. |
| [36] | 田晓明, 颜立红, 向光锋, 等. 植物4香豆酸: 辅酶A连接酶研究进展 [J]. 生物技术通报, 2017, 33(4): 19-26. |
| Tian XM, Yan LH, Xiang GF, et al. Research progress on 4-coumarate: coenzyme a ligase (4CL) in plants [J]. Biotechnol Bull, 2017, 33(4): 19-26. | |
| [37] | 王浩哲, 钱海珍, 尤红艳, 等. 平原红猕猴桃花青苷积累规律及合成代谢关键基因发掘 [J]. 果树学报, 2024, 41(8): 1534-1545. |
| Wang HZ, Qian HZ, You HY, et al. Anthocyanin accumulation and related anabolic key genes in fruit of Pingyuanhong kiwifruit (Actinidia chinensis) [J]. J Fruit Sci, 2024, 41(8): 1534-1545. | |
| [38] | 母洪娜, 孙陶泽, 徐晨, 等. 桂花(Osmanthus fragrans Lour.)4 -香豆酸辅酶A连接酶(4CL)基因克隆与表达分析 [J]. 分子植物育种, 2016, 14(3): 536-541. |
| Mu HN, Sun TZ, Xu C, et al. Gene clone and expression analysis of 4-coumarate-Co a ligase in sweet Osmanthus (Osmanthus fragrans Lour.) [J]. Mol Plant Breed, 2016, 14(3): 536-541. | |
| [39] | Endler A, Martens S, Wellmann F, et al. Unusually divergent 4-coumarate: CoA-ligases from Ruta graveolens L [J]. Plant Mol Biol, 2008, 67(4): 335-346. |
| [40] | 唐亚琴. 油橄榄黄酮和木质素合成相关酶基因4CL的克隆及表达分析 [D]. 雅安: 四川农业大学, 2019. |
| Tang YQ. Cloning and characterization of the flavonoid and lignin synthesis related gene 4CL in olive [D]. Ya’an: Sichuan Agricultural University, 2019. | |
| [41] | Lee SH, Park YJ, Park SU, et al. Expression of genes related to phenylpropanoid biosynthesis in different organs of Ixeris dentata var. albiflora [J]. Molecules, 2017, 22(6): 901. |
| [42] | 潘翔. 毛白杨4CL基因启动子及APX的结构与功能研究 [D]. 北京: 北京林业大学, 2013. |
| Pan X. Study on the structure and function of the 4CL gene promoters and APX in Populus tomentosa [D]. Beijing: Beijing Forestry University, 2013. | |
| [43] | Laoué J, Fernandez C, Ormeño E. Plant flavonoids in mediterranean species: a focus on flavonols as protective metabolites under climate stress [J]. Plants, 2022, 11(2): 172. |
| [44] | Wang JX, Wang X, Ma B, et al. SoNAC72-SoMYB44/SobHLH130 module contributes to flower color fading via regulating anthocyanin biosynthesis by directly binding to the SoUFGT1 promoter in lilac (Syringa oblata) [J]. Hortic Res, 2025, 12(3): uhae326. |
| [45] | 刘淑华, 臧丹丹, 孙燕, 等. 花青素生物合成途径及关键酶研究进展 [J]. 土壤与作物, 2022, 11(3): 336-346. |
| Liu SH, Zang DD, Sun Y, et al. Research advances on biosynthesis pathway of anthocyanins and relevant key enzymes [J]. Soil Crop, 2022, 11(3): 336-346. | |
| [46] | 刘国元, 方威, 余春梅, 等. 花青素调控植物花色的研究进展 [J]. 安徽农业科学, 2021, 49(3): 1-4, 9. |
| Liu GY, Fang W, Yu CM, et al. Research progress on anthocyanins regulated plant flower color [J]. J Anhui Agric Sci, 2021, 49(3): 1-4, 9. | |
| [47] | 彭贞贞, 钟传飞, 王宝刚, 等. ‘红颜’草莓果实成熟过程中花色苷积累及合成途径基因表达的分析 [J]. 食品工业科技, 2023, 44(14): 346-354. |
| Peng ZZ, Zhong CF, Wang BG, et al. Analysis of anthocyanin accumulation and gene expression of anthocyanin synthesis pathway during fruit ripening of ‘benihoppe’ strawberry [J]. Sci Technol Food Ind, 2023, 44(14): 346-354. |
| [1] | 陈静欢, 房国楠, 朱文豪, 叶广继, 苏旺, 贺苗苗, 杨生龙, 周云. 马铃薯种质资源淀粉表征及相关基因表达分析[J]. 生物技术通报, 2026, 42(1): 170-183. |
| [2] | 张永艳, 郭思健, 李晶, 郝思怡, 李瑞得, 刘嘉鹏, 程春振. 蓝莓花青素相关VcGSTF19基因的克隆及功能研究[J]. 生物技术通报, 2025, 41(9): 139-146. |
| [3] | 陈强, 于璎霏, 张颖, 张冲. 茉莉酸甲酯对薄皮甜瓜‘绿宝石’采后冷害的调控[J]. 生物技术通报, 2025, 41(9): 105-114. |
| [4] | 徐小萍, 杨成龙, 和兴, 郭文杰, 吴健, 方少忠. 百合LoAPS1克隆及其在休眠解除过程的功能分析[J]. 生物技术通报, 2025, 41(9): 195-206. |
| [5] | 董向向, 缪百灵, 许贺娟, 陈娟娟, 李亮杰, 龚守富, 朱庆松. 森林草莓FveBBX20基因的生物信息学分析及开花调控功能[J]. 生物技术通报, 2025, 41(9): 115-123. |
| [6] | 程雪, 付颖, 柴晓娇, 王红艳, 邓欣. 谷子LHC基因家族鉴定及非生物胁迫表达分析[J]. 生物技术通报, 2025, 41(8): 102-114. |
| [7] | 李开杰, 吴瑶, 李丹丹. 红花CtbHLH128基因克隆及调控干旱胁迫应答功能研究[J]. 生物技术通报, 2025, 41(8): 234-241. |
| [8] | 任睿斌, 司二静, 万广有, 汪军成, 姚立蓉, 张宏, 马小乐, 李葆春, 王化俊, 孟亚雄. 大麦条纹病菌GH17基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 146-154. |
| [9] | 康琴, 汪霞, 谌明洋, 徐静天, 陈诗兰, 廖平杨, 许文志, 吴卫, 徐东北. 薄荷UV-B受体基因McUVR8的克隆与表达分析[J]. 生物技术通报, 2025, 41(8): 255-266. |
| [10] | 曾丹, 黄园, 王健, 张艳, 刘庆霞, 谷荣辉, 孙庆文, 陈宏宇. 铁皮石斛bZIP转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 197-210. |
| [11] | 韩燚, 侯昌林, 唐露, 孙璐, 谢晓东, 梁晨, 陈小强. 大麦HvERECTA基因的克隆及功能分析[J]. 生物技术通报, 2025, 41(7): 106-116. |
| [12] | 郭秀娟, 冯宇, 吴瑞香, 王利琴, 杨建春. Ca2+处理对胡麻种子萌发影响的转录组分析[J]. 生物技术通报, 2025, 41(7): 139-149. |
| [13] | 魏雨佳, 李岩, 康语涵, 弓晓楠, 杜敏, 涂岚, 石鹏, 于子涵, 孙彦, 张昆. 白颖苔草CrMYB4基因的克隆和表达分析[J]. 生物技术通报, 2025, 41(7): 248-260. |
| [14] | 许慧珍, SHANTWANA Ghimire, RAJU Kharel, 岳云, 司怀军, 唐勋. 马铃薯SUMO E3连接酶基因家族分析及StSIZ1基因的克隆与表达模式分析[J]. 生物技术通报, 2025, 41(6): 119-129. |
| [15] | 裴景琪, 赵梦然, 黄晨阳, 邬向丽. 一个影响糙皮侧耳生长发育的功能基因的发现与验证[J]. 生物技术通报, 2025, 41(6): 327-334. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||