








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 51-66.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0863
刘欢1,2( ), 郭发旭3, 赵晓燕2, 黄龙雨2, 王健1, 周国民4,5,6(
), 郭发旭3, 赵晓燕2, 黄龙雨2, 王健1, 周国民4,5,6( ), 张建华1,2(
), 张建华1,2( )
)
收稿日期:2025-08-09
出版日期:2026-01-26
发布日期:2026-02-04
通讯作者:
张建华,男,博士,研究员,研究方向 :计算机视觉;E-mail: zhangjianhua@caas.cn;作者简介:刘欢,男,硕士研究生,研究方向 :数据分析;E-mail: liuhuan01@139.com
基金资助:
LIU Huan1,2( ), GUO Fa-xu3, ZHAO Xiao-yan2, HUANG Long-yu2, WANG Jian1, ZHOU Guo-min4,5,6(
), GUO Fa-xu3, ZHAO Xiao-yan2, HUANG Long-yu2, WANG Jian1, ZHOU Guo-min4,5,6( ), ZHANG Jian-hua1,2(
), ZHANG Jian-hua1,2( )
)
Received:2025-08-09
Published:2026-01-26
Online:2026-02-04
摘要:
DNA设计是根据特定功能需求定向构建与优化基因组功能元件,已成为合成生物学与精准育种等前沿领域的关键核心技术。传统设计方法受限于对复杂调控网络认知不足及序列搜索空间巨大等瓶颈,难以实现高效、精准的序列创新。近年来,人工智能技术,特别是深度生成模型与预测模型的融合应用,正深刻重塑DNA设计的理论基础与技术范式。该范式通过学习海量组学数据中蕴含的“调控语法”,能够在超长基因组序列背景下实现高分辨率的功能预测、多模态信息整合与条件可控的序列生成。本文系统综述了人工智能在DNA设计中的前沿进展,重点阐述了深度生成与预测模型在启动子、增强子等多层级调控元件设计、序列优化及作物育种等领域的关键技术路径与应用实例。通过构建“设计—预测—优化—验证”的智能化闭环,人工智能不仅显著提升了复杂功能元件的设计效率与准确性,更催生了性能超越天然元件的“超天然”序列。展望未来,随着人工智能与合成生物学、实验自动化等领域的深度融合,DNA设计有望实现从智能化设计到高通量实验验证的完整闭环,从而加速生命科学基础研究与现代农业育种等领域的创新突破。
刘欢, 郭发旭, 赵晓燕, 黄龙雨, 王健, 周国民, 张建华. 人工智能在DNA设计中的研究进展[J]. 生物技术通报, 2026, 42(1): 51-66.
LIU Huan, GUO Fa-xu, ZHAO Xiao-yan, HUANG Long-yu, WANG Jian, ZHOU Guo-min, ZHANG Jian-hua. Advances in Artificial Intelligence for DNA Design[J]. Biotechnology Bulletin, 2026, 42(1): 51-66.
| 模型 Model | 架构类型 Architecture type | 主要用途 Primary applications | 特点 Features | 优势 Advantages | 局限 Limitations |
|---|---|---|---|---|---|
| Enformer | Transformer | 功能预测 | 长程依赖 | 泛化性强 | 数据需求量大 |
| DNABERT | Transformer | 序列特征提取 | k-mer嵌入 | 泛化能力好 | 解释性有限 |
| Evo/Evo2 | Transformer | 序列设计/预测 | 跨模态泛化生 | 零样本迁移 | 复杂度较高 |
| GAN | 对抗生成网络 | 序列设计/预测 | 成创新序列 | 功能多样 | 训练稳定性差 |
| VAE | 自编码器 | 序列优化 | 潜在空间优化 | 控制性强 | 泛化较难 |
| Diffusion | 扩散模型 | 序列精准生成 | 稳定性强 | 可控性强 | 计算资源需求高 |
表1 DNA设计领域常规及最新模型介绍
Table 1 Introduction of conventional and cutting-edge models in the field of DNA design
| 模型 Model | 架构类型 Architecture type | 主要用途 Primary applications | 特点 Features | 优势 Advantages | 局限 Limitations |
|---|---|---|---|---|---|
| Enformer | Transformer | 功能预测 | 长程依赖 | 泛化性强 | 数据需求量大 |
| DNABERT | Transformer | 序列特征提取 | k-mer嵌入 | 泛化能力好 | 解释性有限 |
| Evo/Evo2 | Transformer | 序列设计/预测 | 跨模态泛化生 | 零样本迁移 | 复杂度较高 |
| GAN | 对抗生成网络 | 序列设计/预测 | 成创新序列 | 功能多样 | 训练稳定性差 |
| VAE | 自编码器 | 序列优化 | 潜在空间优化 | 控制性强 | 泛化较难 |
| Diffusion | 扩散模型 | 序列精准生成 | 稳定性强 | 可控性强 | 计算资源需求高 |
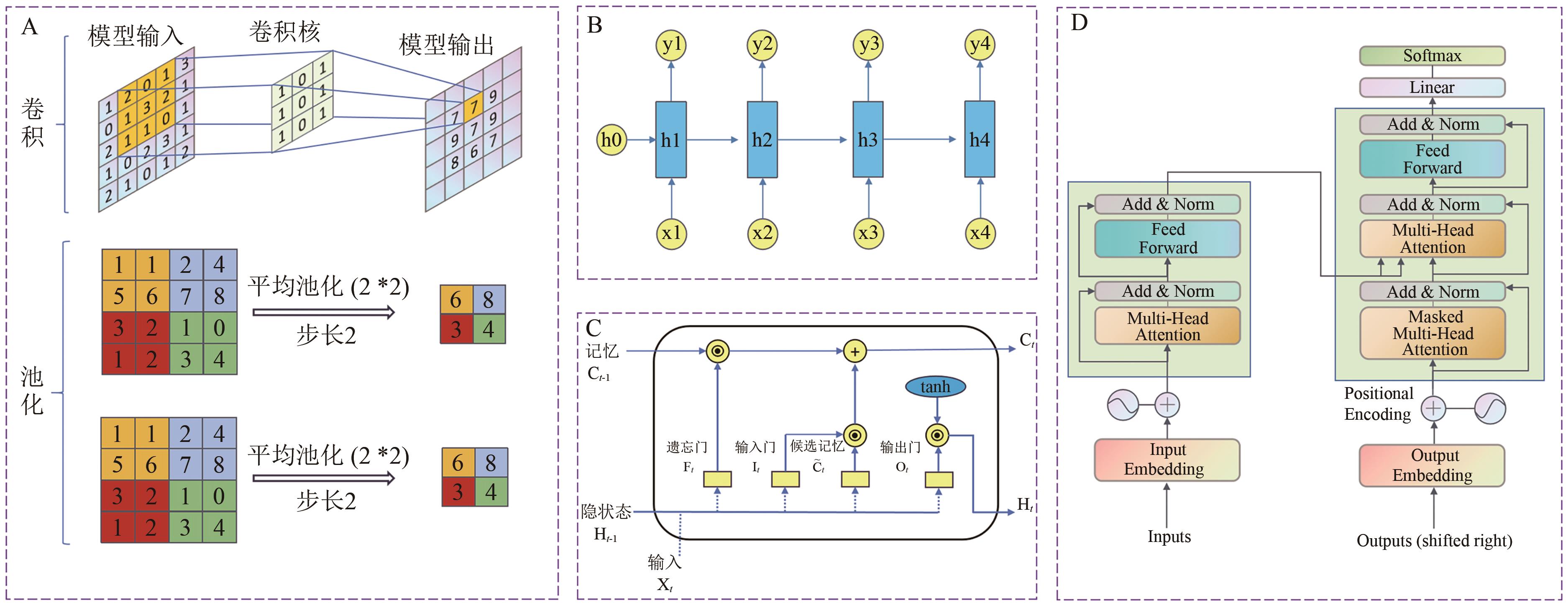
图2 预测模型中不同模型的架构,包括卷积神经网络CNN(A), 循环神经网络RNN(B), 长短时记忆网络LSTM(C)以及Transformer(D)
Fig. 2 Architectures of different predictive models, including (A) a Convolutional Neural Network (CNN), (B) a Recurrent Neural Network (RNN), (C) a Long Short-Term Memory (LSTM) network, and (D) a Transformer
数据库/资源 Database/Resource | 内容简介 Description | 类型/范围 Type/Scope | 用途 Applications |
|---|---|---|---|
| GENCODE | 人类注释基因组 | 注释序列 | 基因注释 |
| INSD | 核酸数据集 | 序列库 | 通用序列建模 |
| EPDnew | 启动子信息 | 启动子功能 | 功能预测 |
| EnhancerAtlas | 增强子注释 | 增强子库 | 功能研究 |
| MethBank | 甲基化数据 | 表观遗传 | 调控分析 |
| UCSC Browser | 多组学数据 | 组学集成 | 多组学分析 |
| GEO | 转录组表观组 | 组学数据库 | 转录组分析 |
| ENCODE | 功能元件注释数据 | 多组学 | 功能元件注释 |
| Roadmap | 表观遗传组 | 表观组 | 功能研究 |
| GTEx | 组织表达图谱 | 转录组 | 表达分析 |
| Ensembl | 多物种注释 | 基因组注释 | 跨物种建模 |
| RNAcentral | ncRNA库 | 非编码RNA | 序列分析 |
| GreeNC | 植物ncRNA | 植物数据库 | 功能元件设计 |
| DBHR | 多组学整合 | 综合组学 | 功能研究 |
| UniProt | 蛋白功能库 | 蛋白注释 | 功能预测 |
| MPRA | 高通量实验 | 功能平台 | 功能筛选 |
表2 DNA设计领域中所使用的数据以及数据来源和用途
Table 2 Data used in the field of DNA design, including their sources and applications
数据库/资源 Database/Resource | 内容简介 Description | 类型/范围 Type/Scope | 用途 Applications |
|---|---|---|---|
| GENCODE | 人类注释基因组 | 注释序列 | 基因注释 |
| INSD | 核酸数据集 | 序列库 | 通用序列建模 |
| EPDnew | 启动子信息 | 启动子功能 | 功能预测 |
| EnhancerAtlas | 增强子注释 | 增强子库 | 功能研究 |
| MethBank | 甲基化数据 | 表观遗传 | 调控分析 |
| UCSC Browser | 多组学数据 | 组学集成 | 多组学分析 |
| GEO | 转录组表观组 | 组学数据库 | 转录组分析 |
| ENCODE | 功能元件注释数据 | 多组学 | 功能元件注释 |
| Roadmap | 表观遗传组 | 表观组 | 功能研究 |
| GTEx | 组织表达图谱 | 转录组 | 表达分析 |
| Ensembl | 多物种注释 | 基因组注释 | 跨物种建模 |
| RNAcentral | ncRNA库 | 非编码RNA | 序列分析 |
| GreeNC | 植物ncRNA | 植物数据库 | 功能元件设计 |
| DBHR | 多组学整合 | 综合组学 | 功能研究 |
| UniProt | 蛋白功能库 | 蛋白注释 | 功能预测 |
| MPRA | 高通量实验 | 功能平台 | 功能筛选 |
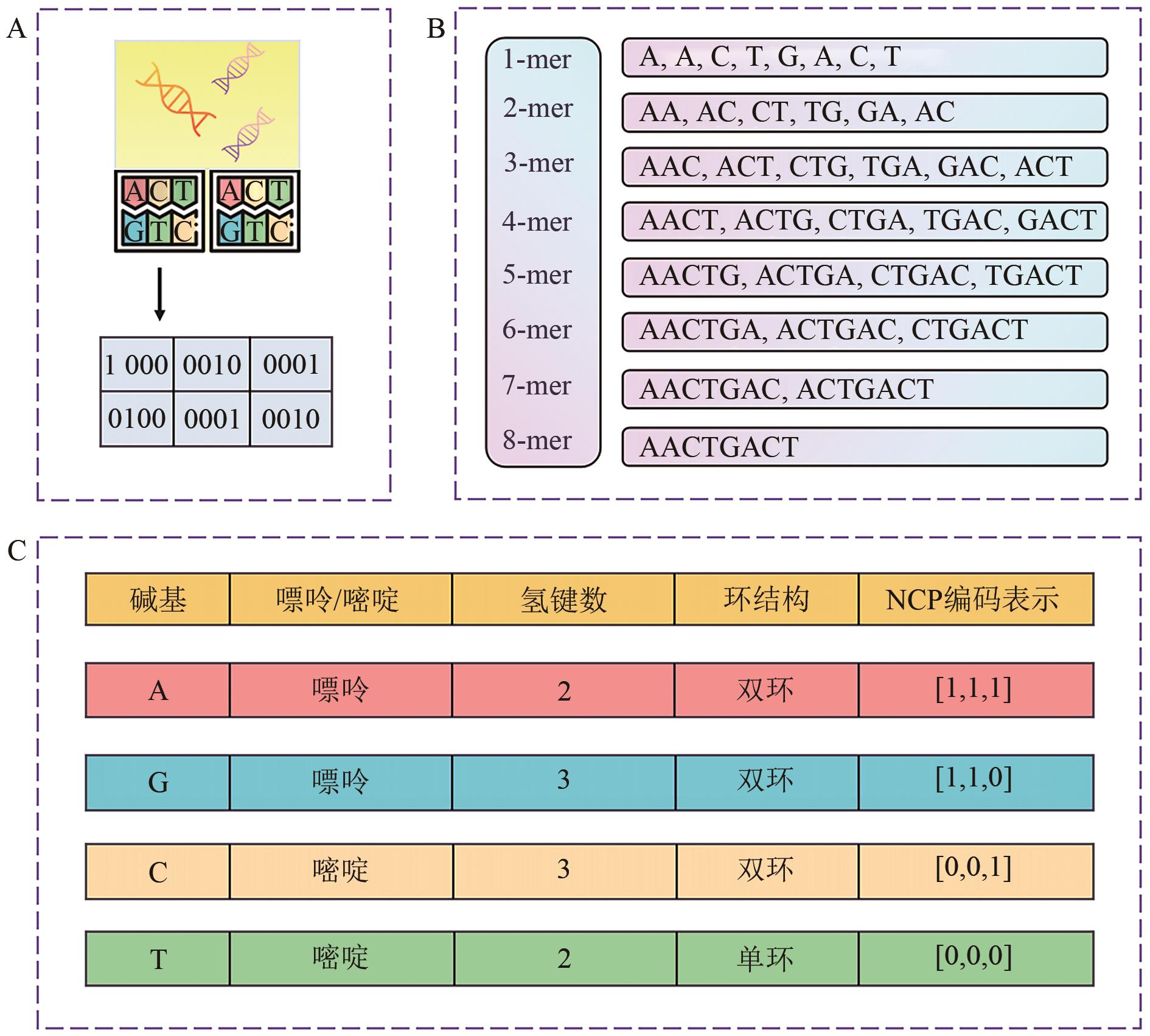
图3 DNA序列的编码方法, 包括独热编码(A), k-mer编码(B), 核苷酸化学性质编码(C)
Fig. 3 DNA sequence encoding methods, including (A) one-hot encoding, (B) k-mer encoding, and (C) nucleotide physicochemical-property encoding
位置 Position | A密度 A density | C密度 C density | G密度 G density | T密度 T density |
|---|---|---|---|---|
| 1 | 1.00 | 0.00 | 0.00 | 0.00 |
| 2 | 0.50 | 0.00 | 0.50 | 0.00 |
| 3 | 0.33 | 0.00 | 0.33 | 0.33 |
| 4 | 0.25 | 0.25 | 0.25 | 0.25 |
| 5 | 0.20 | 0.20 | 0.20 | 0.10 |
| 6 | 0.17 | 0.17 | 0.33 | 0.33 |
| 7 | 0.14 | 0.29 | 0.29 | 0.29 |
| 8 | 0.25 | 0.25 | 0.25 | 0.25 |
表3 ND编码示例和编码后的数值矩阵
Table 3 Example of ND encoding and the resulting numerical matrix
位置 Position | A密度 A density | C密度 C density | G密度 G density | T密度 T density |
|---|---|---|---|---|
| 1 | 1.00 | 0.00 | 0.00 | 0.00 |
| 2 | 0.50 | 0.00 | 0.50 | 0.00 |
| 3 | 0.33 | 0.00 | 0.33 | 0.33 |
| 4 | 0.25 | 0.25 | 0.25 | 0.25 |
| 5 | 0.20 | 0.20 | 0.20 | 0.10 |
| 6 | 0.17 | 0.17 | 0.33 | 0.33 |
| 7 | 0.14 | 0.29 | 0.29 | 0.29 |
| 8 | 0.25 | 0.25 | 0.25 | 0.25 |
| [1] | Eraslan G, Avsec Ž, Gagneur J, et al. Deep learning: new computational modelling techniques for genomics [J]. Nat Rev Genet, 2019, 20(7): 389-403. |
| [2] | Kelley DR, Reshef YA, Bileschi M, et al. Sequential regulatory activity prediction across chromosomes with convolutional neural networks [J]. Genome Res, 2018, 28(5): 739-750. |
| [3] | Agarwal V, Shendure J. Predicting mRNA abundance directly from genomic sequence using deep convolutional neural networks [J]. Cell Rep, 2020, 31(7): 107663. |
| [4] | Zhang PC, Wang HC, Xu HW, et al. Deep flanking sequence engineering for efficient promoter design using DeepSEED [J]. Nat Commun, 2023, 14: 6309. |
| [5] | Wu MR, Nissim L, Stupp D, et al. A high-throughput screening and computation platform for identifying synthetic promoters with enhanced cell-state specificity (SPECS) [J]. Nat Commun, 2019, 10: 2880. |
| [6] | Yu TC, Liu WL, Brinck MS, et al. Multiplexed characterization of rationally designed promoter architectures deconstructs combinatorial logic for IPTG-inducible systems [J]. Nat Commun, 2021, 12: 325. |
| [7] | Ji YR, Zhou ZH, Liu H, et al. DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome [J]. Bioinformatics, 2021, 37(15): 2112-2120. |
| [8] | Linder J, Bogard N, Rosenberg AB, et al. A generative neural network for maximizing fitness and diversity of synthetic DNA and protein sequences [J]. Cell Syst, 2020, 11(1): 49-62.e16. |
| [9] | Zhou ZH, Ji YR, Li WJ, et al. DNABERT-2: Efficient foundation model and benchmark for multi-species genomes [J]. arXiv.org. 2023. DOI: 10.48550/arxiv.2306.15006 . |
| [10] | Avsec Ž, Weilert M, Shrikumar A, et al. Base-resolution models of transcription-factor binding reveal soft motif syntax [J]. Nat Genet, 2021, 53(3): 354-366. |
| [11] | Wang Y, Wang HC, Wei L, et al. Synthetic promoter design in Escherichia coli based on a deep generative network [J]. Nucleic Acids Res, 2020, 48(12): 6403-6412. |
| [12] | Killoran N, Lee LJ, Delong A, et al. Generating and designing DNA with deep generative models [J]. arXiv.org. 2017. DOI: 10.48550/arxiv.1712.06148 . |
| [13] | Gupta A, Zou J. Feedback GAN for DNA optimizes protein functions [J]. Nat Mach Intell, 2019, 1(2): 105-111. |
| [14] | Wang XL, Xu KJ, Tan YM, et al. Deep learning-assisted design of novel promoters in Escherichia coli [J]. Adv Genet, 2023, 4(4): 2300184. |
| [15] | Alipanahi B, Delong A, Weirauch MT, et al. Predicting the sequence specificities of DNA-and RNA-binding proteins by deep learning [J]. Nat Biotechnol, 2015, 33(8): 831-838. |
| [16] | Wang XL, Xu KJ, Huang ZS, et al. Accelerating promoter identification and design by deep learning [J]. Trends Biotechnol, 2025. |
| [17] | Barbadilla-Martínez L, Klaassen N, van Steensel B, et al. Predicting gene expression from DNA sequence using deep learning models [J]. Nat Rev Genet, 2025, 26(10): 666-680. |
| [18] | Quang D, Xie XH. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences [J]. Nucleic Acids Res, 2016, 44(11): e107. |
| [19] | Jores T, Tonnies J, Wrightsman T, et al. Synthetic promoter designs enabled by a comprehensive analysis of plant core promoters [J]. Nat Plants, 2021, 7(6): 842-855. |
| [20] | Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model [J]. Nat Meth, 2015, 12(10): 931-934. |
| [21] | Kelley DR, Snoek J, Rinn JL. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks [J]. Genome Res, 2016, 26(7): 990-999. |
| [22] | Li ZH, Zhang YY, Peng B, et al. A novel interpretable deep learning-based computational framework designed synthetic enhancers with broad cross-species activity [J]. Nucleic Acids Res, 2024, 52(21): 13447-13468. |
| [23] | Yin C, Castillo-Hair S, Byeon GW, et al. Iterative deep learning design of human enhancers exploits condensed sequence grammar to achieve cell-type specificity [J]. Cell Syst, 2025, 16(7): 101302. |
| [24] | Vaishnav ED, de Boer CG, Molinet J, et al. The evolution, evolvability and engineering of gene regulatory DNA [J]. Nature, 2022, 603(7901): 455-463. |
| [25] | Fu HG, Liang YB, Zhong XQ, et al. Codon optimization with deep learning to enhance protein expression [J]. Sci Rep, 2020, 10: 17617. |
| [26] | Yang G, Chen YJ, Guo QH, et al. Leveraging pre-trained AI models for robust promoter sequence design in synthetic biology [J]. Swwlxb, 2025, 11: 1. |
| [27] | Fallahpour A, Gureghian V, Filion GJ, et al. CodonTransformer: a multispecies codon optimizer using context-aware neural networks [J]. Nat Commun, 2025, 16: 3205. |
| [28] | Zrimec J, Fu XZ, Muhammad AS, et al. Controlling gene expression with deep generative design of regulatory DNA [J]. Nat Commun, 2022, 13: 5099. |
| [29] | de Almeida BP, Schaub C, Pagani M, et al. Targeted design of synthetic enhancers for selected tissues in the Drosophila embryo. [J]. Nature, 2024, 626(7997): 207-211. |
| [30] | Avdeyev P, Shi CL, Tan YH, et al. Dirichlet diffusion score model for biological sequence generation [J]. arXiv.org. 2023. DOI: 10.48550/arxiv.2305.10699 . |
| [31] | de Almeida BP, Reiter F, Pagani M, et al. DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers [J]. Nat Genet, 2022, 54(5): 613-624. |
| [32] | Avsec Ž, Agarwal V, Visentin D, et al. Effective gene expression prediction from sequence by integrating long-range interactions [J]. Nat Meth, 2021, 18(10): 1196-1203. |
| [33] | Yang Y, Lee JH, Poindexter MR, et al. Rational design and testing of abiotic stress-inducible synthetic promoters from poplar cis-regulatory elements [J]. Plant Biotechnol J, 2021, 19(7): 1354-1369. |
| [34] | Jain R, Jain A, Mauro E, et al. ICOR: improving codon optimization with recurrent neural networks [J]. BMC Bioinform, 2023, 24(1): 132. |
| [35] | Lei X, Wang X, Chen GL, et al. Combining diffusion and transformer models for enhanced promoter synthesis and strength prediction in deep learning [J]. mSystems, 2025, 10(4) |
| [36] | Kelley DR. Cross-species regulatory sequence activity prediction [J]. PLoS Comput Biol, 2020, 16(7): e1008050. |
| [37] | Li JQ, Zhang PC, Xi X, et al. Modeling and designing enhancers by introducing and harnessing transcription factor binding units [J]. Nat Commun, 2025, 16: 1469. |
| [38] | Gasperini M, Tome JM, Shendure J. Towards a comprehensive catalogue of validated and target-linked human enhancers [J]. Nat Rev Genet, 2020, 21(5): 292-310. |
| [39] | Zhou J, Theesfeld CL, Yao K, et al. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk [J]. Nat Genet, 2018, 50(8): 1171-1179. |
| [40] | Friedman RZ, Ramu A, Lichtarge S, et al. Active learning of enhancers and silencers in the developing neural retina [J]. Cell Syst, 2025, 16(1): 101163. |
| [41] | Karbalayghareh A, Sahin M, Leslie CS. Chromatin interaction-aware gene regulatory modeling with graph attention networks [J]. Genome Res, 2022, 32(7): 1290-1304. |
| [42] | Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, et al. Predicting splicing from primary sequence with deep learning [J]. Cell, 2019, 176(3): 535-548.e24. |
| [43] | Sample PJ, Wang B, Reid DW, et al. Human 5' UTR design and variant effect prediction from a massively parallel translation assay [J]. Nat Biotechnol, 2019, 37(7): 803-809. |
| [44] | Bogard N, Linder J, Rosenberg AB, et al. A deep neural network for predicting and engineering alternative polyadenylation [J]. Cell, 2019, 178(1): 91-106.e23. |
| [45] | Lee NK, Tang ZQ, Toneyan S, et al. EvoAug: improving generalization and interpretability of genomic deep neural networks with evolution-inspired data augmentations [J]. Genome Biol, 2023, 24(1): 105. |
| [46] | Cherednichenko O, Poptsova M. Data augmentation with generative models improves detection of Non-B DNA structures [J]. Comput Biol Med, 2025, 184: 109440. |
| [47] | Davidi D, et al. Regulatory DNA sequence design with reinforcement learning [DB/OL]. arXiv preprint: 2503.07981. |
| [48] | Jaganathan K, Ersaro N, Novakovsky G, et al. Predicting expression-altering promoter mutations with deep learning [J]. Science, 2025, 389(6760): eads7373. |
| [49] | Dalla-Torre H, Gonzalez L, Mendoza-Revilla J, et al. Nucleotide Transformer: building and evaluating robust foundation models for human genomics [J]. Nat Meth, 2025, 22(2): 287-297. |
| [50] | 张冀东, 王志晗, 刘博, 等. 深度学习在生物序列分析领域的应用进展 [J]. 北京工业大学学报, 2022, 48(8): 878-887. |
| Zhang JD, Wang ZH, Liu B, et al. Progress in the applications of deep learning in biological sequence analysis [J]. J Beijing Univ Technol, 2022, 48(8): 878-887. |
| [1] | 蔡如凤, 杨宇轩, 于基正, 李佳楠. 人工智能重塑蛋白质工程:从结构解析到合成生物学的算法革命[J]. 生物技术通报, 2025, 41(8): 1-10. |
| [2] | 王辉, 范灵熙, 孙纪录, 王苑, 伍宁丰, 田健, 关菲菲. 基于蛋白智能模型提升溶菌酶RPL187的热稳定性[J]. 生物技术通报, 2025, 41(7): 336-346. |
| [3] | 郭发旭, 冯全, 张建华, 周焕斌, 杨森, 王健, 周国民. 人工智能驱动的酶改造与设计研究进展[J]. 生物技术通报, 2025, 41(12): 50-65. |
| [4] | 何远, 牟强, 和玉兵, 赵晓燕, 王健, 周国民, 张建华. 基于人工智能的蛋白质挖掘与设计研究进展[J]. 生物技术通报, 2025, 41(10): 143-155. |
| [5] | 王欣, 徐一亿, 徐扬, 徐辰武. 作物全基因组选择育种技术研究进展[J]. 生物技术通报, 2024, 40(3): 1-13. |
| [6] | 纪宏超, 李正艳. 基于质谱的未知次生代谢物结构解析研究进展与展望[J]. 生物技术通报, 2024, 40(10): 76-85. |
| [7] | 彭乙冬;. 英国OROS System公司利用人工智能加速了单克隆抗体的纯化[J]. , 1988, 0(09): 15-15. |
| [8] | 王秀璋;. 美国Focus Tech公司开始人工智能和生物技术结合的保健服务试验[J]. , 1988, 0(02): 22-22. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||