








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 42-50.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0608
收稿日期:2025-06-11
出版日期:2026-01-26
发布日期:2026-02-04
通讯作者:
康晓龙,男,教授,博士生导师,研究方向 :动物基因组与分子育种;E-mail: kangxl9527@126.com作者简介:李慧慧,女,硕士研究生,研究方向 :动物遗传育种与繁殖;E-mail: lihuihuitx@126.com
基金资助:
LI Hui-hui( ), WANG Shu-jie, KANG Xiao-long(
), WANG Shu-jie, KANG Xiao-long( )
)
Received:2025-06-11
Published:2026-01-26
Online:2026-02-04
摘要:
RNA特异性腺苷脱氨酶1(ADAR1)是RNA编辑的关键酶之一,能催化双链RNA(dsRNA)分子中的腺苷(A)转化为肌苷(I),在免疫调节、炎症反应等生理过程中发挥重要作用。ADAR1的免疫调控作用体现在双重机制:其一,通过RNA编辑修饰dsRNA结构,使其无法激活模式识别受体及下游效应蛋白,从而维持自身免疫耐受;其二,通过非编辑依赖性方式与RIG-I、MDA5等模式识别受体互作,间接调控NF-κB、IRF等信号分子的激活,进而影响促炎细胞因子的释放。在炎症反应中,ADAR1通过多条路径发挥抗炎效应。一方面,其可编辑病毒或自身dsRNA,降低dsRNA对RIG-I、MDA5的激活能力,从而抑制抗病毒炎症的过度激活;另一方面,ADAR1与PKR直接相互作用,抑制eIF2α磷酸化,以平衡细胞存活与炎症性死亡;同时,ADAR1还能通过编辑内源性dsRNA,阻断OAS-RNase L通路的异常RNA降解,避免由此触发的自身炎症;此外,ADAR1与ZBP1竞争性结合Z-RNA,抑制RIPK3-MLKL介导的程序性坏死,从而减轻由坏死引发的炎症反应。这些机制为开发针对炎症性疾病的治疗策略提供了理论基础,然而,上述研究多集中于人类和小鼠模型,其在家养动物炎症性疾病中的调控作用尚未明确。本文综述了ADAR1在炎症和免疫方面的研究,归纳了其重要调控机制,以期为揭示家养动物的炎症性疾病如奶牛乳房炎等的发病机理及开发有效防治策略奠定基础。
李慧慧, 王书节, 康晓龙. ADAR1在固有免疫和炎症中的调控作用[J]. 生物技术通报, 2026, 42(1): 42-50.
LI Hui-hui, WANG Shu-jie, KANG Xiao-long. The Role of RNA-specific Adenosine Deaminase 1 in Innate Immunity and Inflammation[J]. Biotechnology Bulletin, 2026, 42(1): 42-50.
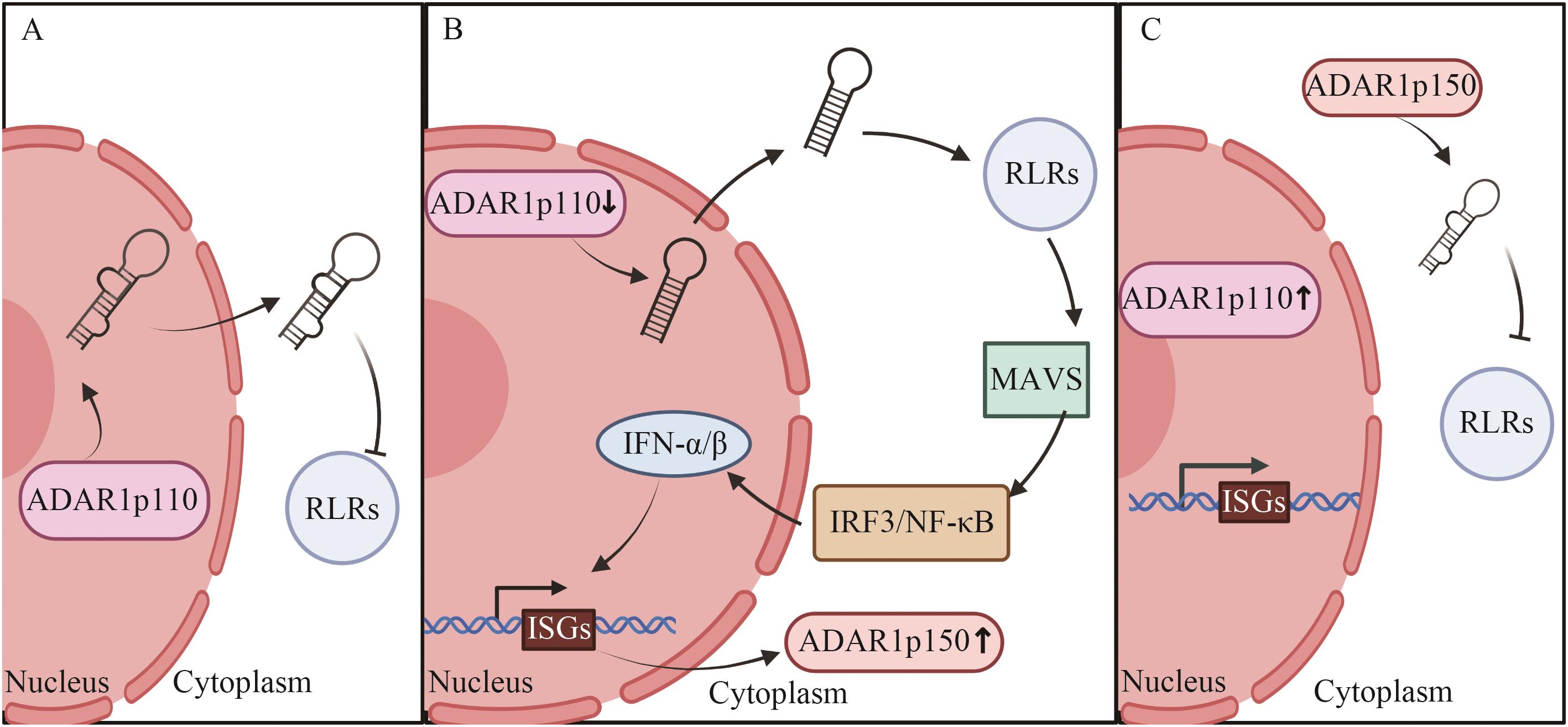
图1 病毒感染时ADAR1两个亚型的表达与作用A:未感染病毒时;B:感染病毒前期;C:感染病毒后期。IFN-α/β:Ⅰ型IFN
Fig. 1 Expression and function of two subtypes of ADAR1 in virus infectionA: When the virus is not infected. B: The early stage of viral infection. C: The late stage of viral infection. IFN-α/β: Type I IFN
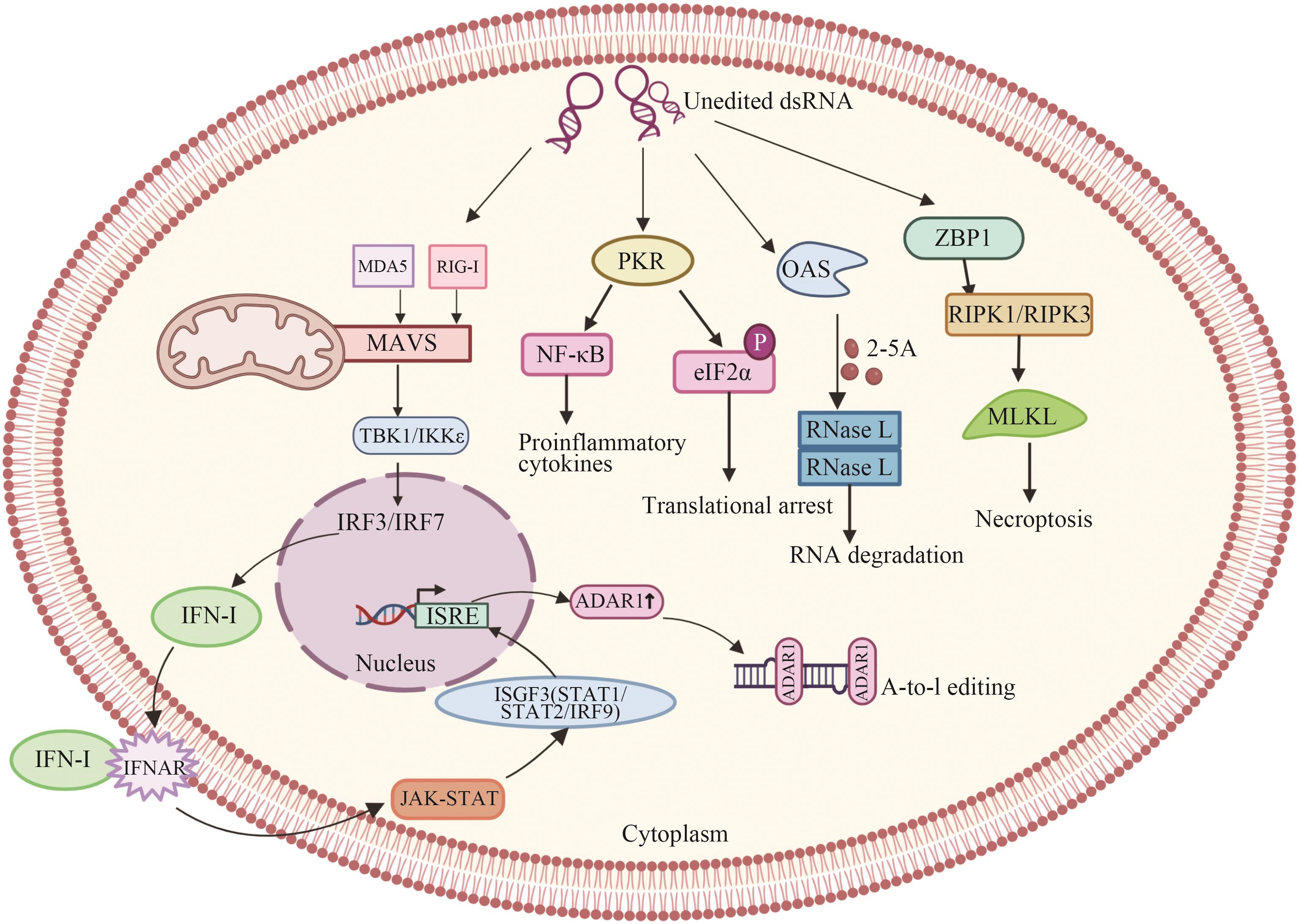
图2 ADAR1在固有免疫中的作用IRF3/IRF7:转录因子;NF-κB:核因子-κB;IFN-Ⅰ:I型IFN;IFNAR:IFN的受体;ISGs:干扰素刺激基因;ISGF3:ISGF3(interferon-stimulated gene factor 3)是一种转录因子复合物,由STAT1、STAT2和IRF9组成
Fig. 2 Role of ADAR1 in innate immunityIRF3/IRF7: Transcription factor; NF-κB: nuclear factor-κB; IFN-Ⅰ: type I IFN; IFNAR: receptor for IFN; ISGs, interferon stimulating gene; ISGF3: interferon-stimulated gene factor 3 (ISGF3) is a transcription factor complex composed of STAT1, STAT2, and IRF9
| [1] | Song YL, Yang WB, Fu Q, et al. irCLASH reveals RNA substrates recognized by human ADARs [J]. Nat Struct Mol Biol, 2020, 27(4): 351-362. |
| [2] | Zhong YH, Zhong X, Qiao LJ, et al. Zα domain proteins mediate the immune response [J]. Front Immunol, 2023, 14: 1241694. |
| [3] | Bahn JH, Ahn J, Lin XZ, et al. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways [J]. Nat Commun, 2015, 6: 6355. |
| [4] | Song B, Shiromoto Y, Minakuchi M, et al. The role of RNA editing enzyme ADAR1 in human disease [J]. Wires RNA, 2022, 13(1): e1665. |
| [5] | Kleinova R, Rajendra V, Leuchtenberger AF, et al. The ADAR1 editome reveals drivers of editing-specificity for ADAR1-isoforms [J]. Nucleic Acids Res, 2023, 51(9): 4191-4207. |
| [6] | Ashley CN, Broni E, Miller WA. ADAR family proteins: a structural review [J]. Curr Issues Mol Biol, 2024, 46(5): 3919-3945. |
| [7] | Matthews MM, Thomas JM, Zheng YX, et al. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity [J]. Nat Struct Mol Biol, 2016, 23(5): 426-433. |
| [8] | Fisher AJ, Beal PA. Structural perspectives on adenosine to inosine RNA editing by ADARs [J]. Mol Ther Nucleic Acids, 2024, 35(3): 102284. |
| [9] | Quin J, Sedmík J, Vukić D, et al. ADAR RNA modifications, the epitranscriptome and innate immunity [J]. Trends Biochem Sci, 2021, 46(9): 758-771. |
| [10] | Song CZ, Sakurai M, Shiromoto Y, et al. Functions of the RNA editing enzyme ADAR1 and their relevance to human diseases [J]. Genes, 2016, 7(12): 129. |
| [11] | Xu L-D, Öhman M. ADAR1 editing and its role in cancer [J]. Genes, 2019, 10(1): 12. |
| [12] | Liddicoat BJ, Piskol R, Chalk AM, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself [J]. Science, 2015, 349(6252): 1115-1120. |
| [13] | Samuel CE. Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses [J]. J Biol Chem, 2019, 294(5): 1710-1720. |
| [14] | Yuan J, Xu L, Bao HJ, et al. Biological roles of A-to-I editing: implications in innate immunity, cell death, and cancer immunotherapy [J]. J Exp Clin Cancer Res, 2023, 42(1): 149. |
| [15] | Adamczak D, Fornalik M, Małkiewicz A, et al. ADAR1 expression in different cancer cell lines and its change under heat shock [J]. J Appl Genet, 2024: 149. |
| [16] | Szymczak F, Cohen-Fultheim R, Thomaidou S, et al. ADAR1-dependent editing regulates human β cell transcriptome diversity during inflammation [J]. Front Endocrinol, 2022, 13: 1058345. |
| [17] | Valentine A, Bosart K, Bush W, et al. Identification and characterization of ADAR1 mutations and changes in gene expression in human cancers [J]. Cancer Genet, 2024, 288/289: 82-91. |
| [18] | Mboukou A, Rajendra V, Messmer S, et al. Dimerization of ADAR1 modulates site-specificity of RNA editing [J]. Nat Commun, 2024, 15: 10051. |
| [19] | Roth SH, Danan-Gotthold M, Ben-Izhak M, et al. Increased RNA editing may provide a source for autoantigens in systemic lupus erythematosus [J]. Cell Rep, 2018, 23(1): 50-57. |
| [20] | Nakahama T, Kawahara Y. The RNA-editing enzyme ADAR1: a regulatory hub that tunes multiple dsRNA-sensing pathways [J]. Int Immunol, 2023, 35(3): 123-133. |
| [21] | Bazak L, Haviv A, Barak M, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes [J]. Genome Res, 2014, 24(3): 365-376. |
| [22] | Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs [J]. Nat Rev Mol Cell Biol, 2016, 17(2): 83-96. |
| [23] | Mu JQ, Wu C, Xu KM, et al. Conformational reorganization and phase separation drive hyper-editing of ADR-2-ADBP-1 complex [J]. Nucleic Acids Res, 2025, 53(5): gkaf148. |
| [24] | Uzonyi A, Nir R, Shliefer O, et al. Deciphering the principles of the RNA editing code via large-scale systematic probing [J]. Mol Cell, 2021, 81(11): 2374-2387.e3. |
| [25] | Rehwinkel J, Mehdipour P. ADAR1: from basic mechanisms to inhibitors [J]. Trends Cell Biol, 2025, 35(1): 59-73. |
| [26] | Deffit SN, Hundley HA. To edit or not to edit: regulation of ADAR editing specificity and efficiency [J]. Wiley Interdiscip Rev RNA, 2016, 7(1): 113-127. |
| [27] | Sharma M, Wagh P, Shinde T, et al. Exploring the role of pattern recognition receptors as immunostimulatory molecules [J]. Immunity Inflam & Disease, 2025, 13(5): e70150. |
| [28] | Chen RC, Zou J, Chen JW, et al. Pattern recognition receptors: function, regulation and therapeutic potential [J]. Signal Transduct Target Ther, 2025, 10(1): 216. |
| [29] | Lamers MM, van den Hoogen BG, Haagmans BL. ADAR1: “editor-in-chief” of cytoplasmic innate immunity [J]. Front Immunol, 2019, 10: 1763. |
| [30] | Sikorska J, Wyss DF. Recent developments in understanding RIG-I’s activation and oligomerization [J]. Sci Prog, 2024, 107(3): 368504241265182. |
| [31] | Liu SQ, Cai X, Wu JX, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation [J]. Science, 2015, 347(6227): aaa2630. |
| [32] | Ivashkiv LB, Donlin LT. Regulation of type I interferon responses [J]. Nat Rev Immunol, 2014, 14(1): 36-49. |
| [33] | Sampaio NG, Chauveau L, Hertzog J, et al. The RNA sensor MDA5 detects SARS-CoV-2 infection [J]. Sci Rep, 2021, 11(1): 13638. |
| [34] | Yang SY, Deng P, Zhu ZW, et al. Adenosine deaminase acting on RNA 1 limits RIG-I RNA detection and suppresses IFN production responding to viral and endogenous RNAs [J]. J Immunol, 2014, 193(7): 3436-3445. |
| [35] | Jiang YF, Zhang HY, Wang J, et al. Exploiting RIG-I-like receptor pathway for cancer immunotherapy [J]. J Hematol Oncol, 2023, 16(1): 8. |
| [36] | Dempsey LA. ADAR1 regulates mda5-MAVS [J]. Nat Immunol, 2016, 17(1): 47. |
| [37] | Shiromoto Y, Sakurai M, Minakuchi M, et al. ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells [J]. Nat Commun, 2021, 12(1): 1654. |
| [38] | Kim JI, Nakahama T, Yamasaki R, et al. RNA editing at a limited number of sites is sufficient to prevent MDA5 activation in the mouse brain [J]. PLoS Genet, 2021, 17(5): e1009516. |
| [39] | Deng P, Khan A, Jacobson D, et al. Adar RNA editing-dependent and-independent effects are required for brain and innate immune functions in Drosophila [J]. Nat Commun, 2020, 11(1): 1580. |
| [40] | Gal-Ben-Ari S, Barrera I, Ehrlich M, et al. PKR: a kinase to remember [J]. Front Mol Neurosci, 2019, 11: 480. |
| [41] | Hu SB, Heraud-Farlow J, Sun T, et al. ADAR1p150 prevents MDA5 and PKR activation via distinct mechanisms to avert fatal autoinflammation [J]. bioRxiv, 2023: 2023.01.25.525475. |
| [42] | Gannon HS, Zou T, Kiessling MK, et al. Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells [J]. Nat Commun, 2018, 9(1): 5450. |
| [43] | Pestal K, Funk CC, Snyder JM, et al. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development [J]. Immunity, 2015, 43(5): 933-944. |
| [44] | Sinigaglia K, Cherian A, Du QP, et al. An ADAR1 dsRBD3-PKR kinase domain interaction on dsRNA inhibits PKR activation [J]. Cell Rep, 2024, 43(8): 114618. |
| [45] | Clerzius G, Gélinas JF, Daher A, et al. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication [J]. J Virol, 2009, 83(19): 10119-10128. |
| [46] | Toth AM, Li ZQ, Cattaneo R, et al. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR [J]. J Biol Chem, 2009, 284(43): 29350-29356. |
| [47] | Gélinas JF, Clerzius G, Shaw E, et al. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase [J]. J Virol, 2011, 85(17): 8460-8466. |
| [48] | Wang YB, Holleufer A, Gad HH, et al. Length dependent activation of OAS proteins by dsRNA [J]. Cytokine, 2020, 126: 154867. |
| [49] | Daou S, Talukdar M, Tang JL, et al. A phenolic small molecule inhibitor of RNase L prevents cell death from ADAR1 deficiency [J]. Proc Natl Acad Sci USA, 2020, 117(40): 24802-24812. |
| [50] | Li YZ, Banerjee S, Goldstein SA, et al. Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line [J]. eLife, 2017, 6: e25687. |
| [51] | Jiao HP, Wachsmuth L, Kumari S, et al. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation [J]. Nature, 2020, 580(7803): 391-395. |
| [52] | Devos M, Tanghe G, Gilbert B, et al. Sensing of endogenous nucleic acids by ZBP1 induces keratinocyte necroptosis and skin inflammation [J]. J Exp Med, 2020, 217(7): e20191913. |
| [53] | Kesavardhana S, Subbarao Malireddi RK, Burton AR, et al. The Zα2 domain of ZBP1 is a molecular switch regulating influenza-induced PANoptosis and perinatal lethality during development [J]. J Biol Chem, 2020, 295(24): 8325-8330. |
| [54] | Chen XY, Dai YH, Wan XX, et al. ZBP1-mediated necroptosis: mechanisms and therapeutic implications [J]. Molecules, 2022, 28(1): 52. |
| [55] | Zhan JH, Wang JS, Liang YQ, et al. Apoptosis dysfunction: unravelling the interplay between ZBP1 activation and viral invasion in innate immune responses [J]. Cell Commun Signal, 2024, 22(1): 149. |
| [56] | Karki R, Sundaram B, Sharma BR, et al. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis [J]. Cell Rep, 2021, 37(3): 109858. |
| [57] | Jiao HP, Wachsmuth L, Wolf S, et al. ADAR1 averts fatal type I interferon induction by ZBP1 [J]. Nature, 2022, 607(7920): 776-783. |
| [58] | de Reuver R, Verdonck S, Dierick E, et al. ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation [J]. Nature, 2022, 607(7920): 784-789. |
| [59] | Zhang YG, Zhang JY, Xue YC. ADAR1: a mast regulator of aging and immunity [J]. Signal Transduct Target Ther, 2023, 8(1): 7. |
| [1] | 王一凡, 朱鸿亮. 植物PPR蛋白的功能及其作用机制[J]. 生物技术通报, 2025, 41(6): 27-37. |
| [2] | 虎喜敏, 周冉, 王正兴, 李宇航, 罗仍卓么, 王兴平. 干扰lncRNA RNF5-AS1对奶牛乳腺上皮细胞炎症反应的影响[J]. 生物技术通报, 2025, 41(5): 333-342. |
| [3] | 李新颖, 孙晶, 吕若彤, 任亚娟, 罗蕾, 艾鹏飞, 王雁伟. PPR蛋白调控叶绿体RNA编辑分子机制研究进展[J]. 生物技术通报, 2025, 41(10): 32-42. |
| [4] | 杨微, 关海峰, 任欣慧, 彭金菊, 陈志宝. 虾青素对黄曲霉毒素B1诱导肝损伤的缓解作用及机制[J]. 生物技术通报, 2025, 41(10): 334-342. |
| [5] | 雷棋怡, 徐杨, 李鹏飞. 脆弱拟杆菌六型分泌系统对肠道屏障的影响及机制[J]. 生物技术通报, 2024, 40(3): 286-295. |
| [6] | 马文澳, 杨微, 李迎春, 朱彦彬, 陈志宝, 刘娜. 桦褐孔菌乙醇提取物对脂多糖诱导肠损伤的缓解作用及机制[J]. 生物技术通报, 2024, 40(12): 299-308. |
| [7] | 吴永娜, 滕文龙, 张磊, 王德富, 牛颜冰. 连翘叶茶对大鼠肝硬化的影响及其机理研究[J]. 生物技术通报, 2024, 40(11): 285-295. |
| [8] | 龙佳佳, 刘玮玮, 范新浩, 黎旺长, 杨小淦, 唐中林. 基于大规模RNA-seq数据绘制猪RNA编辑图谱的研究[J]. 生物技术通报, 2024, 40(10): 288-295. |
| [9] | 焦帅, 付域泽, 崔凯, 张吉贤, 王杰, 毕研亮, 刁其玉, 张建新, 张乃锋. 短小芽孢杆菌对羔羊肠道炎症和屏障功能的影响[J]. 生物技术通报, 2024, 40(1): 344-352. |
| [10] | 陈彩萍, 任昊, 龙腾飞, 何冰, 鲁兆祥, 孙坚. 大肠杆菌Nissle 1917对炎症性肠病治疗作用的研究进展[J]. 生物技术通报, 2023, 39(6): 109-118. |
| [11] | 陈英, 王艺磊, 邹鹏飞. 大黄鱼TRAF6的克隆及表达分析[J]. 生物技术通报, 2022, 38(8): 233-243. |
| [12] | 朱琳, 鲜凤君, 张倩楠, 胡骏. RNA编辑的研究进展[J]. 生物技术通报, 2022, 38(1): 1-14. |
| [13] | 李凯晴, 李莹, 王艺磊, 邹鹏飞. 受体相互作用蛋白的功能及在硬骨鱼类中的研究进展[J]. 生物技术通报, 2021, 37(5): 197-211. |
| [14] | 张立兴, 王丽娜, 康广博, 黄鹤. 多组学分析在炎症性肠病中的应用与研究进展[J]. 生物技术通报, 2021, 37(1): 155-167. |
| [15] | 陈敏洁, 唐桂月, 洪香娜, 郝沛, 江静, 李轩. 基于CRISPR-Cas13家族的RNA编辑系统及其最新 进展[J]. 生物技术通报, 2020, 36(3): 1-8. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||