








生物技术通报 ›› 2021, Vol. 37 ›› Issue (4): 96-106.doi: 10.13560/j.cnki.biotech.bull.1985.2020-0887
收稿日期:2020-07-16
出版日期:2021-04-26
发布日期:2021-05-13
作者简介:张瑶心,女,博士,讲师,研究方向:微生物生理生化;E-mail:基金资助:
ZHANG Yao-xin( ), WANG Liang-jie, ZHENG Wen, XU Han-qin, ZHENG Lian, ZHONG Jing(
), WANG Liang-jie, ZHENG Wen, XU Han-qin, ZHENG Lian, ZHONG Jing( )
)
Received:2020-07-16
Published:2021-04-26
Online:2021-05-13
摘要:
从湖北武汉地区虾壳堆积土壤中筛选得到一株分泌胞外几丁质酶菌株ZWW8,通过16S rDNA序列比对发现其属于无色杆菌属;通过碳源、氮源、温度、培养基pH、培养时间的单因素优化实验获得ZWW8菌株的最优发酵条件,该菌株在添加0.5%几丁质胶体,0.5% NH4Cl,初始pH 7.0的培养基中,于37℃培养2 d后胞外上清酶活力可达到33.9 U/L,是优化前酶活力的8倍;通过检测ZWW8菌株分泌胞外几丁质酶的酶学性质发现,其最适反应pH为5,最适反应温度为50℃。Ni 2+和Ca 2+能提高几丁质酶活性,而Hg 2+、Cu 2+和Zn 2+能明显抑制酶活。此外,几丁质酶还能在高盐条件维持活性和稳定性,并特异性结合几丁质。通过几丁质亲和层析纯化的胞外几丁质酶,在透析除去尿素后仍然具有活性。ZWW8菌株的几丁质酶具有独特的酶学特性,使其能作为高效稳定的生物催化剂,为实现在工业、农业和医学方面的应用潜能,以及几丁质资源的开发和利用奠定基础。
张瑶心, 王亮节, 郑文, 徐汉琴, 郑恋, 钟静. 产几丁质酶的无色杆菌ZWW8的发酵产酶及酶学性质研究[J]. 生物技术通报, 2021, 37(4): 96-106.
ZHANG Yao-xin, WANG Liang-jie, ZHENG Wen, XU Han-qin, ZHENG Lian, ZHONG Jing. Study on Enzyme Production of a Chitinase-producing Strain Achromobacter sp. ZWW8 by Fermentation and Its Enzymatic Characterization[J]. Biotechnology Bulletin, 2021, 37(4): 96-106.
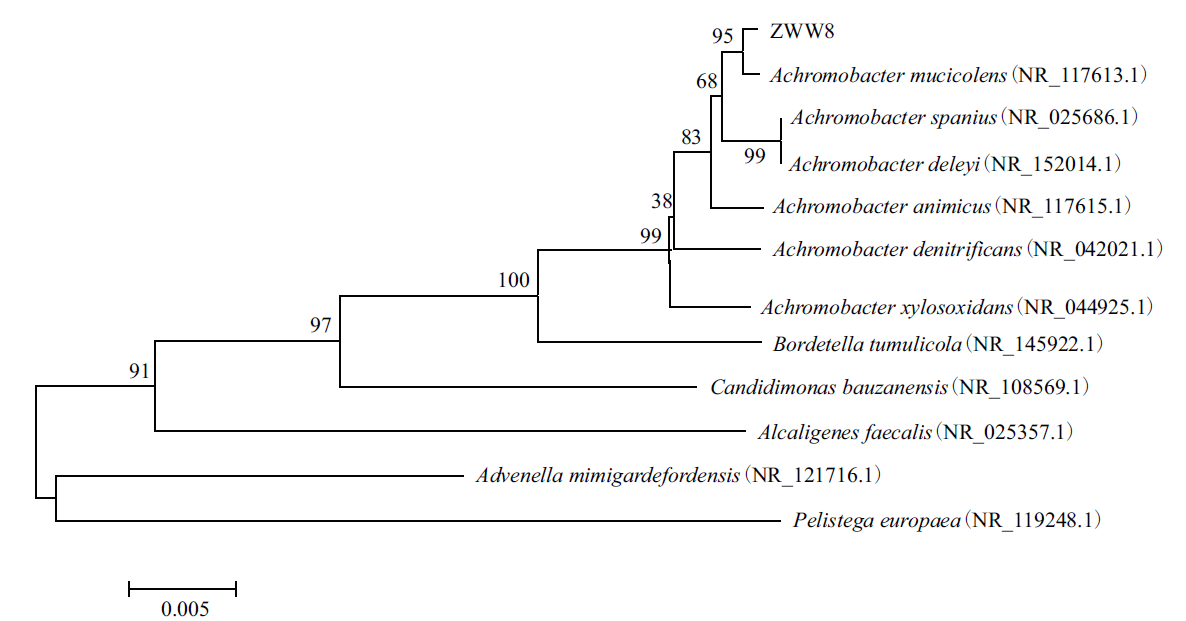
图1 基于16S rDNA序列构建的ZWW8菌株的系统进化树 括号中的序号代表菌株的GenBank 登录号;分支点上的数字代表计算1 000 次聚类到一起的几率;标尺刻度代表0.5%的序列
Figure 1 Phylogenetic tree of strain ZWW8 based on 16S rDNA sequences Numbers in parentheses refer to the sequence’ accession number in GenBank. The number at each branch point is the percentage supported by bootstrap. Bar: 0.5% sequence divergence
| 碳源Carbon source | 酶活Enzyme activity/(U·L-1) |
|---|---|
| 葡萄糖 Glucose | 0.2±0.8 |
| 胶体几丁质 Colloidal chitin | 20.0±1.9 |
| 粉末几丁质 Chitin powder | 4.2±0.3 |
| 胶体壳聚糖 Colloidal chitosan | 0.0±0.1 |
| 水溶性壳聚糖 Soluble chitosan | 0.2±0.1 |
| N-乙酰氨基葡萄糖 N-Acetyl-D-glucosamine | 0.4±0.6 |
表1 不同碳源对ZWW8菌株发酵产几丁质酶的影响
Table 1 Effect of different carbon sources on chitinase production by Achromobacter sp. ZWW8
| 碳源Carbon source | 酶活Enzyme activity/(U·L-1) |
|---|---|
| 葡萄糖 Glucose | 0.2±0.8 |
| 胶体几丁质 Colloidal chitin | 20.0±1.9 |
| 粉末几丁质 Chitin powder | 4.2±0.3 |
| 胶体壳聚糖 Colloidal chitosan | 0.0±0.1 |
| 水溶性壳聚糖 Soluble chitosan | 0.2±0.1 |
| N-乙酰氨基葡萄糖 N-Acetyl-D-glucosamine | 0.4±0.6 |
| 氮源Nitrogen source | 酶活Enzyme activity(U·L-1) |
|---|---|
| 氯化铵 Ammonium chloride | 20.7±1.3 |
| 硫酸铵 Ammonium sulfate | 17.8±1.5 |
| 硝酸钠 Sodium nitrate | 6.4±1.9 |
| 尿素 Urea | 13.3±1.0 |
| 蛋白胨 Peptone | 12.1±1.8 |
| 硝酸铵Ammonium nitrate | 19.2±0.2 |
表2 不同氮源对ZWW8菌株发酵产几丁质酶的影响
Table2 Effect of different nitrogen sources on chitinase production by Achromobacter sp. ZWW8
| 氮源Nitrogen source | 酶活Enzyme activity(U·L-1) |
|---|---|
| 氯化铵 Ammonium chloride | 20.7±1.3 |
| 硫酸铵 Ammonium sulfate | 17.8±1.5 |
| 硝酸钠 Sodium nitrate | 6.4±1.9 |
| 尿素 Urea | 13.3±1.0 |
| 蛋白胨 Peptone | 12.1±1.8 |
| 硝酸铵Ammonium nitrate | 19.2±0.2 |
| 金属离子Metal ion | 相对酶活Relative activity/% |
|---|---|
| Hg2+ | 3.1±3.3 |
| Zn2+ | 84.4±8.5 |
| Mg2+ | 96.1±7.1 |
| Ca2+ | 103.9±9.3 |
| Ag+ | 99.2±6.8 |
| Cu2+ | 75.9±0.1 |
| Ni2+ | 114.7±10.3 |
表3 金属离子对ZWW8菌株几丁质酶活性的影响
Table 3 Effect of metal ions on chitinase activity by strain ZWW8
| 金属离子Metal ion | 相对酶活Relative activity/% |
|---|---|
| Hg2+ | 3.1±3.3 |
| Zn2+ | 84.4±8.5 |
| Mg2+ | 96.1±7.1 |
| Ca2+ | 103.9±9.3 |
| Ag+ | 99.2±6.8 |
| Cu2+ | 75.9±0.1 |
| Ni2+ | 114.7±10.3 |

图11 几丁质亲和层析纯化ZWW8菌株几丁质酶的SDS-PAGE检测图 M:低分子量蛋白Marker;1:粗酶液;2:上样流出液;3、4:洗涤液;5:洗脱液
Fig.11 SDS-PAGE analysis of strain ZWW8 chitinase purified by chitin affinity chromatography M: Low molecular weight marker. 1: Crude chitinase. 2: Effluent. 3,4: Scrub solution, 5: Purified chitinase
| [1] |
Kurita K. Chemistry and application of chitin and chitosan[J]. Polymer Degradation and Stability, 1998,59(1):117-120.
doi: 10.1016/S0141-3910(97)00160-2 URL |
| [2] | 欧阳石文, 冯兰香, 赵开军. 几丁质酶的三级结构和催化机制[J]. 生命的化学, 2001,21(2):131-133. |
| Ou-Yang SW, Feng LX, Zhao KJ. The tertiary structure and catalytic mechanism of chitinase[J]. Chemistry of Life, 2001,21(2):131-133. | |
| [3] |
Bhattacharya D, Nagpure A, Gupta RK. Bacterial chitinases:properties and potential[J]. Critical Reviews in Biotechnology, 2007,27(1):21-28.
doi: 10.1080/07388550601168223 URL |
| [4] |
Nagpure A, Bharti , Gupta RK. Chitinases:in agriculture and human healthcare[J]. Critical Reviews in Biotechnology, 2013,34(3):215-232.
doi: 10.3109/07388551.2013.790874 URL |
| [5] | Dahiya N, Tewari R, Hoondal GS. Biotechnological aspects of chitinolytic enzymes:a review[J]. Applied Microbiology & Biotechnology, 2006,71(6):773-782. |
| [6] | 郭玉莲. 微生物几丁质酶及其在植物病害防治中的作用[J]. 中国农学通报, 2005(1):283-285. |
| Guo Y. Chitinase and its effects on plant disease prevention[J]. Chinese Agricultural Science Bulletin, 2005(1):283-285. | |
| [7] | 黄乾生, 谢晓兰, 陈清西. 几丁质酶的结构特征与功能[J]. 厦门大学学报:自然科学版, 2008,47(z2). 232-235. |
| Huang QS, Xie XL, Chen QX. The structure and function of chitinase[J]. Journal of Xiamen University:Natural Science, 2008,47(z2). 232-235 . | |
| [8] | 冯金荣, 惠丰立, 文祯中. 真菌几丁质酶及其在植物真菌病害防治中的作用[J]. 河南农业科学, 2006(8):83-86. |
| Feng JR, Hui FL, Wen ZZ. Fungal chitinase and its role in the control of plant fungal diseases[J]. Journal of Henan Agricultural Sciences, 2006(8):83-86. | |
| [9] | Sakuda S, Isogai A, Makita T, et al. Structures of allosamidins, novel insect chitinase inhibitors, produced by actinomycetes[J]. Journal of the Agricultural Chemical Society of Japan, 2006,51(12):3251-3259. |
| [10] |
Liaqat F, Eltem R. Chitooligosaccharides and their biological activities:A comprehensive review[J]. Carbohydrate Polymers, 2018,184:243-259.
doi: 10.1016/j.carbpol.2017.12.067 URL |
| [11] |
Chigaleĭchik AG, Pirieva DA, Rylkin SS, et al. Biosynjournal of chitinase by Achromobacter liquefaciens[J]. Mikrobiologiia, 1976,45:475.
pmid: 12452 |
| [12] | Takashi T, Hideyuki M. Antibacterial, anti-nematode and/or plant-cell activating composition,chitinolytic microorganisms for producing the same:European, EP0401560[P]. 1993-05-04. |
| [13] | Bholay AD, Bhushan K, Rameez S, et al. Novel chitinolytic potential of Achromobacter denitrificans isolated from fishery waste[J]. IOSR Journal of Pharmacy and Biological Sciences, 2015,10(4):31-36. |
| [14] |
Baker GC, Smith JJ, Cowan DA . Review and re-analysis of domain-specific 16S primers[J]. Journal of Microbiological Methods, 2004,55(3):541-555.
doi: 10.1016/j.mimet.2003.08.009 URL |
| [15] | 张军毅, 朱冰川, 徐超, 等. 基于分子标记的宏基因组16S rRNA基因高变区选择策略[J]. 应用生态学报, 2015,26(11):3545-3553. |
| Zhang JY, Zhu BC, Xu C, et al. Strategy of selecting 16S rRNA hypervariable regions for matagenome-phylogenetic marker genes based analysis[J]. Chinese Journal of Applied Ecology, 2015,26(11):3545-3553. | |
| [16] |
Kim SK, Rajapakse N. Enzymatic production and biological activities of chitosan oligosaccharides(COS):A review[J]. Carbohydrate Polymers, 2005,62(4):357-368.
doi: 10.1016/j.carbpol.2005.08.012 URL |
| [17] |
Dou J, Xu Q, Tan C, et al. Effects of chitosan oligosaccharides on neutrophils from glycogen-induced peritonitis mice model[J]. Carbohydrate Polymers, 2009,75(1):119-124.
doi: 10.1016/j.carbpol.2008.07.005 URL |
| [18] |
Aam BB, Heggset EB, Norberg AL, et al. Production of chitooligosaccharides and their potential applications in medicine[J]. Marine Drugs, 2010,8(5):1482-1517.
doi: 10.3390/md8051482 URL |
| [19] |
Hough DW, Danson MJ. Extremozymes[J]. Current Opinion in Chemical Biology, 1999,3(1):39-46.
pmid: 10021406 |
| [20] |
Karan R, Capes MD, Dassarma S . Function and biotechnology of extremophilic enzymes in low water activity[J]. Aquatic Biosystems, 2012,8(1):1-4.
doi: 10.1186/2046-9063-8-1 URL |
| [21] |
Vaidya R, Shah I, Vyas P, et al. Production of chitinase and its optimization from a novel isolate Alcaligenes xylosoxydans:potential in antifungal biocontrol[J]. World Journal of Microbiology & Biotechnology, 2001,17(7):691-696.
doi: 10.1023/A:1012927116756 URL |
| [22] | Vaidya R, Vyas P, Chhatpar HS. Statistical optimization of medium components for the production of chitinase by Alcaligenes xylosoxydans[J]. Enzyme & Microbial Technology, 2003,33(1):92-96. |
| [23] |
Vaidya R, Roy S, Macmil S, et al. Purification and characterization of chitinase from Alcaligenes xylosoxydans[J]. Biotechnology Letters, 2003,25(9):715-717.
doi: 10.1023/A:1023406630791 URL |
| [24] | 付星, 闫巧娟, 江正强, 等. 高产几丁质酶巴伦葛兹类芽孢杆菌的筛选和发酵条件优化[J]. 微生物学通报, 2015,42(4):625-633. |
| Fu X, Yan QJ, Jiang ZQ, et al. Screening of a high-level chitinase producing strain, Paenibacillus barengoltzii and optimization of its fermentation conditions[J]. Microbiology China, 2015,42(4):625-633. | |
| [25] |
Vyas P, Deshpande MV . Chitinase production by Myrothecium verrucaria and its significance for fungal mycelia degradation[J]. J Gen Appl Microbiol, 1989,35(5):343-350.
doi: 10.2323/jgam.35.343 URL |
| [26] |
Kavitha A, Vijaylakshmi M. Partial purification and antifungal profile of chitinase produced by Streptomyces tendae TK-VL_333[J]. Annals of Microbiology, 2011,61(3):597-603.
doi: 10.1007/s13213-010-0178-1 URL |
| [27] |
Uchiyama T, Katouno F, Nikaidou N, et al. Roles of the exposed aromatic residues in crystalline chitin hydrolysis by chitinase A from Serratia marcescens 2170[J]. Journal of Biological Chemistry, 2001,276(44):41343-41349.
doi: 10.1074/jbc.M103610200 URL |
| [28] |
Orikoshi H, Nakayama S, Miyamoto K, et al. Roles of four chitinases(ChiA, ChiB, ChiC, and ChiD)in the chitin degradation system of marine bacterium Alteromonas sp. strain O-7[J]. Applied and Environmental Microbiology, 2005,71(4):1811-1815.
doi: 10.1128/AEM.71.4.1811-1815.2005 URL |
| [1] | 程亚楠, 张文聪, 周圆, 孙雪, 李玉, 李庆刚. 乳酸乳球菌生产2'-岩藻糖基乳糖的途径构建及发酵培养基优化[J]. 生物技术通报, 2023, 39(9): 84-96. |
| [2] | 韩志阳, 贾子苗, 梁秋菊, 王轲, 唐华丽, 叶兴国, 张双喜. 二套小麦-簇毛麦染色体附加系苗期耐盐性及籽粒硒和叶酸的含量[J]. 生物技术通报, 2023, 39(8): 185-193. |
| [3] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [4] | 车永梅, 郭艳苹, 刘广超, 叶青, 李雅华, 赵方贵, 刘新. 菌株C8和B4的分离鉴定及其耐盐促生效果和机制[J]. 生物技术通报, 2023, 39(5): 276-285. |
| [5] | 赵赛赛, 张小丹, 贾晓妍, 陶大炜, 刘可玉, 宁喜斌. 高产硝酸盐还原酶Staphylococcus simulans ZSJ6的复合诱变选育及其酶学性质研究[J]. 生物技术通报, 2023, 39(4): 103-113. |
| [6] | 杨俊钊, 张新蕊, 赵国柱, 郑菲. 新型GH5家族多结构域纤维素酶的结构与功能研究[J]. 生物技术通报, 2023, 39(4): 71-80. |
| [7] | 王凤婷, 王岩, 孙颖, 崔文婧, 乔凯彬, 潘洪玉, 刘金亮. 耐盐碱土曲霉SYAT-1的分离鉴定及抑制植物病原真菌特性研究[J]. 生物技术通报, 2023, 39(2): 203-210. |
| [8] | 陈奕博, 杨万明, 岳爱琴, 王利祥, 杜维俊, 王敏. 基于SLAF标记的大豆遗传图谱构建及苗期耐盐性QTL定位[J]. 生物技术通报, 2023, 39(2): 70-79. |
| [9] | 车永梅, 刘广超, 郭艳苹, 叶青, 赵方贵, 刘新. 一种耐盐复合菌剂的制备和促生作用研究[J]. 生物技术通报, 2023, 39(11): 217-225. |
| [10] | 张玉娟, 黎冬华, 宫慧慧, 崔新晓, 高春华, 张秀荣, 游均, 赵军胜. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析[J]. 生物技术通报, 2023, 39(11): 308-317. |
| [11] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [12] | 刘佳欣, 张会龙, 邹荣松, 杨秀艳, 朱建峰, 张华新. 不同类型盐生植物适应盐胁迫的生理生长机制及Na+逆向转运研究进展[J]. 生物技术通报, 2023, 39(1): 59-72. |
| [13] | 李霁虹, 荆玉玲, 马桂珍, 郭荣君, 李世东. 无色杆菌77的基因组构成及其趋化和耐药特性[J]. 生物技术通报, 2022, 38(9): 136-146. |
| [14] | 王雨辰, 丁尊丹, 关菲菲, 田健, 刘国安, 伍宁丰. 耐热漆酶ba4基因鉴定与酶学性质分析[J]. 生物技术通报, 2022, 38(8): 252-260. |
| [15] | 赵忠娟, 杨凯, 扈进冬, 魏艳丽, 李玲, 徐维生, 李纪顺. 盐胁迫条件下哈茨木霉ST02对椒样薄荷生长及根区土壤理化性质的影响[J]. 生物技术通报, 2022, 38(7): 224-235. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||