








生物技术通报 ›› 2021, Vol. 37 ›› Issue (10): 120-127.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1497
熊亮斌1,2( ), 孙吉1, 刘显舟1, 屈占国1, 计雨情1, 徐一新1(
), 孙吉1, 刘显舟1, 屈占国1, 计雨情1, 徐一新1( ), 王风清2(
), 王风清2( )
)
收稿日期:2020-12-09
出版日期:2021-10-26
发布日期:2021-11-12
作者简介:熊亮斌,男,助理研究员,研究方向:代谢工程及合成生物学;E-mail: 基金资助:
XIONG Liang-bin1,2( ), SUN Ji1, LIU Xian-zhou1, QU Zhan-guo1, JI Yu-qing1, XU Yi-xin1(
), SUN Ji1, LIU Xian-zhou1, QU Zhan-guo1, JI Yu-qing1, XU Yi-xin1( ), WANG Feng-qing2(
), WANG Feng-qing2( )
)
Received:2020-12-09
Published:2021-10-26
Online:2021-11-12
摘要:
为探究新金色分枝杆菌代谢甾醇积累甾体药物中间体过程的中心代谢调节机制,采用转录组测序结合qRT-PCR技术,对比分析甾药中间体生产菌株中心代谢的关键基因转录水平,测定转化过程的葡萄糖消耗速率。结果显示,糖酵解途径的pfkB、pyk转录下调1.96倍及1.49倍;磷酸戊糖途径的zwf、gntZ转录下调3.57倍及2.43倍;三羧酸循环的citA、icd2和kdg转录分别下调2.5倍、1.78倍及1.92倍。对葡萄糖消耗速率的测定显示,中间体生产菌株的初始代谢速率显著低于野生型菌株。研究结果表明在转化甾醇积累甾药中间体过程中,分枝杆菌的中心代谢途径受到一定程度的抑制性调节。
熊亮斌, 孙吉, 刘显舟, 屈占国, 计雨情, 徐一新, 王风清. 分枝杆菌转化甾醇过程的中心代谢关键基因转录差异分析[J]. 生物技术通报, 2021, 37(10): 120-127.
XIONG Liang-bin, SUN Ji, LIU Xian-zhou, QU Zhan-guo, JI Yu-qing, XU Yi-xin, WANG Feng-qing. Transcription Variations of Key Genes in the Central Metabolism Pathways of the Sterols Transformation in Mycobacterium[J]. Biotechnology Bulletin, 2021, 37(10): 120-127.
| 名称Name | 描述Description | 来源Source |
|---|---|---|
| 菌株 | ||
| Escherichia coli DH5α | 用于质粒转化扩增 | 天根生化 |
| Mycobacterium neoaurum ATCC 25795(Mn) | 野生型菌株 | ATCC |
| MnΔkstD1(MnΔk1) | 在野生型Mn菌株中删除kstD1基因,具有转化甾醇积累9-OHAD的生产能力 | 姚抗等[ |
| 质粒 | ||
| p2NIL | 用于分枝杆菌基因敲除的同源臂构建载体,KanR | Dr. Parish惠赠 |
| p2N-kstD1 | 携带kstD1基因敲除同源臂的p2NIL重组质粒 | 姚抗等[ |
| pGOAL19 | HygR, Pag85-lacZ, Phsp60-sacB, Pac I cassette vector, AmpR | Dr. Parish惠赠 |
| p19-kstD1 | p2NIL携带kstD1基因敲除同源臂及pGOAL19筛选表达框组成的重组质粒,KanR和HygR | 姚抗等[ |
表1 本研究涉及的菌株及质粒
Table 1 Strains and plasmids used in this study
| 名称Name | 描述Description | 来源Source |
|---|---|---|
| 菌株 | ||
| Escherichia coli DH5α | 用于质粒转化扩增 | 天根生化 |
| Mycobacterium neoaurum ATCC 25795(Mn) | 野生型菌株 | ATCC |
| MnΔkstD1(MnΔk1) | 在野生型Mn菌株中删除kstD1基因,具有转化甾醇积累9-OHAD的生产能力 | 姚抗等[ |
| 质粒 | ||
| p2NIL | 用于分枝杆菌基因敲除的同源臂构建载体,KanR | Dr. Parish惠赠 |
| p2N-kstD1 | 携带kstD1基因敲除同源臂的p2NIL重组质粒 | 姚抗等[ |
| pGOAL19 | HygR, Pag85-lacZ, Phsp60-sacB, Pac I cassette vector, AmpR | Dr. Parish惠赠 |
| p19-kstD1 | p2NIL携带kstD1基因敲除同源臂及pGOAL19筛选表达框组成的重组质粒,KanR和HygR | 姚抗等[ |
| 用途 Purpose | 名称Name | 描述Description |
|---|---|---|
| 基因删除引物 Primers for gene deletion | D-kstD1-UF-Hind III | CAGTaagcttCTTCTCAGCCATACGTGGCTCCTA |
| D-kstD1-UR-Pst I | ttaactgcaggtcctgggcagtcatgtagaacac | |
| D-kstD1-DF-Pst I | tacactgcagttgcatctcgctggaaaggcctga | |
| D-kstD1-DR-Kpn I | TATAggtaccCGCGGTCAGCGTTCCGATGAACTT | |
| qRT-PCR分析引物 Primers for qRT-PCR analysis | R-16S rRNA-F | CCTATGTTGCCAGCGGGTTATGC & GCGATTACTAGCGACTCCGACTTCA |
| R-pfkA-F & R | CCAGGCTTGAACGCCGTGATCAG & GATCGGGATCAACACATCGATGC | |
| R-pfkB-F & R | TACCTGCTGCTGCCGACGATCCG & ACCACGTCGGCGTACCAGCTCGT | |
| R-pyk-F & R | GACCACGAAGAGTCCTATCGCCG & ACCAGTCCGACGTTGCCGTCGTC | |
| R-zwf-F & R | CTGATGCCGGCGATCTACGACCT & GTAGAACGCGTGATTGCCGTTGG | |
| R-gntZ-F & R | CTGACCGAAGGCTACGGATTGAT & GCAGCCAGCATCTCGTAACCCTC | |
| R-citA-F & R | GACATCGACGACCTGGTATCGGA & ACATAGGACAGCGCCATCGCCGA | |
| R-icd2-F & R | ATCAAGCTGCCGAACATCAGCGC & TGGCCCATGCTGTGCGGGTGCTT | |
| R-kdg-F & R | CGGCCAAGGCGATGATCGACAAC & CATCGTCTCGCACTTCTTGATGG |
表2 本研究涉及的引物
Table 2 Primers used in this study
| 用途 Purpose | 名称Name | 描述Description |
|---|---|---|
| 基因删除引物 Primers for gene deletion | D-kstD1-UF-Hind III | CAGTaagcttCTTCTCAGCCATACGTGGCTCCTA |
| D-kstD1-UR-Pst I | ttaactgcaggtcctgggcagtcatgtagaacac | |
| D-kstD1-DF-Pst I | tacactgcagttgcatctcgctggaaaggcctga | |
| D-kstD1-DR-Kpn I | TATAggtaccCGCGGTCAGCGTTCCGATGAACTT | |
| qRT-PCR分析引物 Primers for qRT-PCR analysis | R-16S rRNA-F | CCTATGTTGCCAGCGGGTTATGC & GCGATTACTAGCGACTCCGACTTCA |
| R-pfkA-F & R | CCAGGCTTGAACGCCGTGATCAG & GATCGGGATCAACACATCGATGC | |
| R-pfkB-F & R | TACCTGCTGCTGCCGACGATCCG & ACCACGTCGGCGTACCAGCTCGT | |
| R-pyk-F & R | GACCACGAAGAGTCCTATCGCCG & ACCAGTCCGACGTTGCCGTCGTC | |
| R-zwf-F & R | CTGATGCCGGCGATCTACGACCT & GTAGAACGCGTGATTGCCGTTGG | |
| R-gntZ-F & R | CTGACCGAAGGCTACGGATTGAT & GCAGCCAGCATCTCGTAACCCTC | |
| R-citA-F & R | GACATCGACGACCTGGTATCGGA & ACATAGGACAGCGCCATCGCCGA | |
| R-icd2-F & R | ATCAAGCTGCCGAACATCAGCGC & TGGCCCATGCTGTGCGGGTGCTT | |
| R-kdg-F & R | CGGCCAAGGCGATGATCGACAAC & CATCGTCTCGCACTTCTTGATGG |
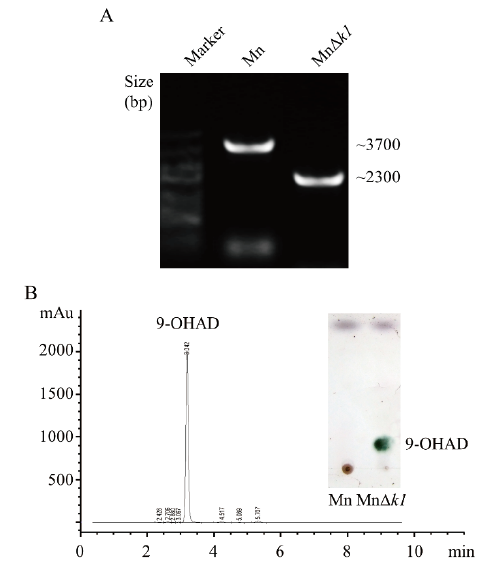
图1 删除kstD1基因的分枝杆菌PCR验证及转化甾醇结果 A:PCR扩增验证结果。Mn:野生型菌株的PCR扩增结果;MnΔk1:缺失kstD1基因菌株的PCR验证结果。B:MnΔk1菌株转化甾醇发酵液的HPLC和TLC分析结果
Fig. 1 Effect of deleting kstD1 on the Mycobacterium PCR verification and biotransformation of sterols A: Evidence for allelic replacement of kstD1 gene. Mn: The wild-type strain. MnΔk1: Deletion of kstD1 in the Mn strain. B: HPLC and TLC analysis results of the sterol biotransformation in MnΔk1 strain
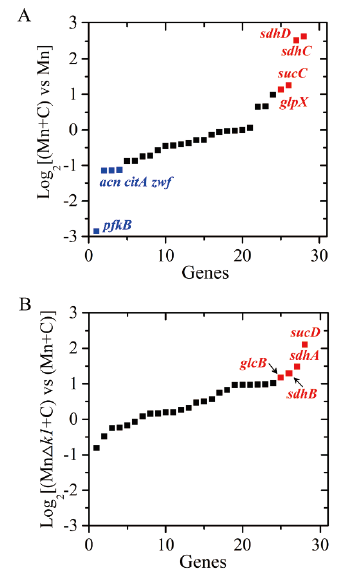
图2 分枝杆菌转化甾醇过程中心代谢相关基因的转录对比分析 A:野生型菌株Mn代谢甾醇的转录变化。B:MnΔk1代谢甾醇过程的转录变化。Mn:野生型菌株接种于不含甾醇的发酵培养液;Mn+C:野生型菌株接种于添加1 g/L甾醇的发酵培养液;MnΔk1+C:9-OHAD初始转化菌株接种于添加1 g/L甾醇的发酵培养液
Fig. 2 Transcriptional comparisons of genes related in central metabolism pathways during the sterol transformation in Mycobacterium A: Transcript profiles of the genes in wild-type Mn strain. B: Transcript profiles of the genes in MnΔk1 strain. Mn: The wild type strain was cultivated in fermentation medium without phytosterol addition. Mn+C: The wild type strain was cultured in fermentation medium with 1 g/L of phytosterol addition. MnΔk1+C: the strain MnΔk1 was cultured in fermentation medium with 1 g/L of phytosterol addition
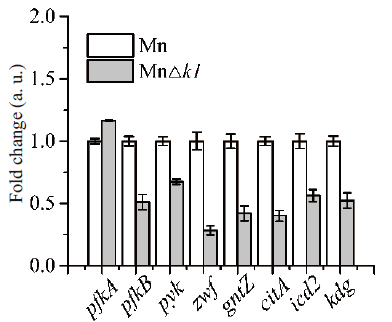
图3 中心代谢关键基因转录的qRT-PCR分析结果 pfkA & pfkB,6-磷酸果糖激酶;pyk,丙酮酸激酶;zwf,葡萄糖-6-磷酸脱氢酶基因;gntZ,6-磷酸葡萄糖酸脱氢酶;citA,柠檬酸合酶;icd2,异柠檬酸脱氢酶;kdg,酮戊二酸脱氢酶
Fig. 3 qRT-PCR analysis of key genes in central metabolism pfkA & pfkB: 6-phosphofructokinase; pyk: pyruvate kinase; zwf: glucose 6-phosphatedehydrogenase; gntZ: 6-phosphogluconate dehydrogenase; citA: citrate synthase; icd2: isocitric dehydrogenase; kdg: oxoglutarate dehydrogenase
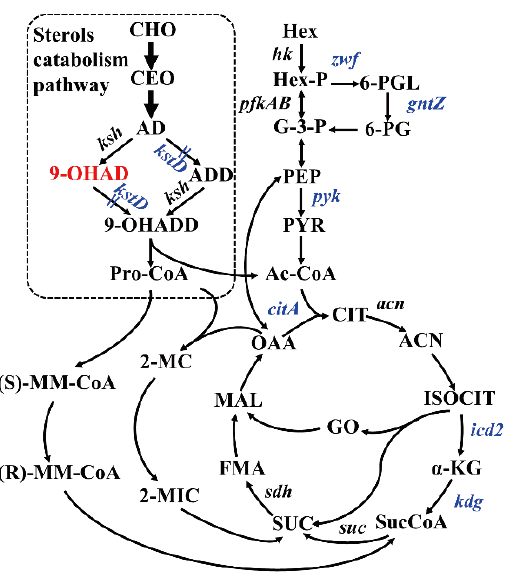
图5 分枝杆菌甾醇代谢与中心代谢途径的关联简图 CHO:胆甾醇;CEO:胆甾酮;AD:雄甾-4-烯-3,17-二酮;ADD:雄甾-1,4-二烯-3,17-二酮;9-OHAD,9α-羟基化-雄甾-4-烯-3,17-二酮;9-OHADD:9α-羟基化-雄甾-1,4-二烯-3,17-二酮;Pro-CoA:丙酰辅酶A;Hex:己糖;Hex-P:磷酸己糖;G-3-P:甘油醛3-磷酸;PEP:磷酸烯醇式丙酮酸;PYR:丙酮酸;Ac-CoA:乙酰辅酶A;(S)-MM-CoA:(S)-甲基丙二酰-CoA;(R)-MM-CoA:(R)-甲基丙二酰-CoA;2-MC:2-甲基柠檬酸;2-MIC:2-甲基异柠檬酸;CIT:柠檬酸;CAN:乌头酸;ISOCIT:异柠檬酸;α-KG:α-酮戊二酸;SucCoA:琥珀酰-CoA;SUC:琥珀酸;FMA:延胡索酸;MAL:苹果酸;OAA:草酰乙酸
Fig. 5 Schematic diagram of sterol metabolism pathway and central metabolism pathway in Mycobacterium CHO: cholesterol; CEO: cholesterone; AD: 4-androstene-3,17-dione; ADD: 1,4-androstadiene-3,17-dione; 9-OHAD: 9α-hydroxy-4-androstene-3,17-dione; 9-OHADD: 9 α-hydroxy-1,4-androstadiene-3,17-dione; Pro-CoA: propionyl-CoA; Hex: hexose; Hex-P: hexose phosphate; G-3-P: glyceraldehyde 3-phosphate; PEP: phosphoenolpyruvate; PYR: pyruvic acid; Ac-CoA: acetyl-CoA; (S)-MM-CoA: (S)-methylmalonyl-CoA; (R)-MM-CoA: (R)-methylmalonyl-CoA; 2-MC: 2-methycitrate; 2-MIC: 2-methylisocitrate; CIT: citrate; ACN: aconitic acid; ISOCIT: isocitrate; α-KG: α-ketoglutarate; SucCoA: succinyl-CoA; SUC: succinate; FMA: fumarate; MAL: malate; OAA: oxaloacetate
| [1] |
Vallée M, Vitiello S, Bellocchio L, et al. Pregnenolone can protect the brain from cannabis intoxication[J]. Science, 2014, 343(6166):94-98.
doi: 10.1126/science.1243985 URL |
| [2] | 熊亮斌. 分枝杆菌甾醇代谢途径的解析及高产甾体医药中间体工程菌株的构建[D]. 上海:华东理工大学, 2017. |
| Xiong LB. Analysis of the sterol metabolic pathway in mycobacteria and the modification of high-yield steroidal pharmaceutical precursors producing strains[D]. Shanghai:East China University of Science and Technology, 2017. | |
| [3] | Donova MV, Dovbnya DV, Sukhodolskaya GV, et al. Microbial conversion of sterol-containing soybean oil production waste[J]. Journal of Chemical Technology and Biotechnology, 2005, 80(1):55-60. |
| [4] |
Yao K, Xu LQ, Wang FQ, et al. Characterization and engineering of 3-ketosteroid-Δ1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3, 17-dione through the catabolism of sterols[J]. Metabolic Engineering, 2014, 24:181-191.
doi: 10.1016/j.ymben.2014.05.005 URL |
| [5] | Liu HH, Xu LQ, Yao K, et al. Characterization and engineering of 3-ketosteroid 9α-hydroxylases in Mycobacterium neoaurum ATCC 25795 for the development of androst-1, 4-diene-3, 17-dione and 9α-hydroxy-androst-4-ene-3, 17-dione producing strains[J]. Applied and Environmental Microbiology, 2018, 84(14):e-02777-02717. |
| [6] |
Xu LQ, Liu YJ, Yao K, et al. Unraveling and engineering the production of 23, 24-bisnorcholenic steroids in sterol metabolism[J]. Scientific Reports, 2016, 6:21928.
doi: 10.1038/srep21928 pmid: 26898409 |
| [7] |
Donova MV, Egorova OV. Microbial steroid transformations:current state and prospects[J]. Applied Microbiology and Biotechnology, 2012, 94(6):1423-1447.
doi: 10.1007/s00253-012-4078-0 URL |
| [8] |
Fernández-Cabezón L, Galán B, García JL. Unravelling a new catabolic pathway of C-19 steroids in Mycobacterium smegmatis[J]. Environmental Microbiology, 2018, 20(5):1815-1827.
doi: 10.1111/1462-2920.14114 pmid: 29611894 |
| [9] |
Wipperman MF, Sampson NS, Thomas ST. Pathogen roid rage:cholesterol utilization by Mycobacterium tuberculosis[J]. Critical Reviews in Biochemistry and Molecular Biology, 2014, 49(4):269-293.
doi: 10.3109/10409238.2014.895700 pmid: 24611808 |
| [10] |
Liu M, Xiong LB, Tao X, et al. Metabolic adaptation of Mycobacterium neoaurum ATCC 25795 in the catabolism of sterols for producing important steroid intermediates[J]. Journal of Agricultural and Food Chemistry, 2018, 66(45):12141-12150.
doi: 10.1021/acs.jafc.8b04777 URL |
| [11] |
Xiong LB, Liu HH, Xu LQ, et al. Role identification and application of SigD in the transformation of soybean phytosterol to 9α-hydroxy-4-androstene-3, 17-dione in Mycobacterium neoaurum[J]. Journal of Agricultural and Food Chemistry, 2017, 65(3):626-631.
doi: 10.1021/acs.jafc.6b05314 URL |
| [12] |
Xiong LB, Sun WJ, Liu YJ, et al. Enhancement of 9α-hydroxy-4-androstene-3, 17-dione production from soybean phytosterols by deficiency of a regulated intramembrane proteolysis metalloprotease in Mycobacterium neoaurum[J]. Journal of Agricultural and Food Chemistry, 2017, 65(48):10520-10525.
doi: 10.1021/acs.jafc.7b03766 URL |
| [13] |
Yao K, Wang FQ, Zhang HC, et al. Identification and engineering of cholesterol oxidases involved in the initial step of sterols catabolism in Mycobacterium neoaurum[J]. Metabolic Engineering, 2013, 15:75-87.
doi: 10.1016/j.ymben.2012.10.005 URL |
| [14] |
Gonzalo-Asensio J, Malaga W, Pawlik A, et al. Evolutionary history of tuberculosis shaped by conserved mutations in the PhoPR virulence regulator[J]. Proceedings of the National Academy of Sciences USA, 2014, 111(31):11491-11496.
doi: 10.1073/pnas.1406693111 URL |
| [15] | Broset E, Martín C, Gonzalo-Asensio J. Evolutionary landscape of the Mycobacterium tuberculosis complex from the viewpoint of PhoPR:implications for virulence regulation and application to vaccine development[J]. Mbio, 2015, 6(5):e01289. |
| [16] |
Xiong LB, Liu HH, Zhao M, et al. Enhancing the bioconversion of phytosterols to steroidal intermediates by the deficiency of kasB in the cell wall synjournal of Mycobacterium neoaurum[J]. Microbial Cell Factories, 2020, 19:80.
doi: 10.1186/s12934-020-01335-y URL |
| [17] |
Xiong LB, Liu HH, Song XW, et al. Improving the biotransformation of phytosterols to 9α-hydroxy-4-androstene-3, 17-dione by deleting embC associated with the assembly of cell envelope in Mycobacterium neoaurum[J]. Journal of Biotechnology, 2020, 323(2020):341-346.
doi: 10.1016/j.jbiotec.2020.09.019 URL |
| [18] |
Baker JJ, Johnson BK, Abramovitch RB. Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources[J]. Molecular Microbiology, 2014, 94(1):56-69.
doi: 10.1111/mmi.2014.94.issue-1 URL |
| [19] |
Xiong LB, Liu HH, Xu LQ, et al. Improving the production of 22-hydroxy-23, 24-bisnorchol-4-ene-3-one from sterols in Mycobacterium neoaurum by increasing cell permeability and modifying multiple genes[J]. Microbial Cell Factories, 2017, 16:89.
doi: 10.1186/s12934-017-0705-x URL |
| [1] | 赵光绪, 杨合同, 邵晓波, 崔志豪, 刘红光, 张杰. 一株高效溶磷产红青霉培养条件优化及其溶磷特性[J]. 生物技术通报, 2023, 39(9): 71-83. |
| [2] | 董聪, 高庆华, 王玥, 罗同阳, 王庆庆. 基于联合策略提高FAD依赖的葡萄糖脱氢酶的酵母表达[J]. 生物技术通报, 2023, 39(6): 316-324. |
| [3] | 张君, 张虹, 张芮, 路国栋, 雍婧姣, 郎思睿, 陈任. 甜菊醇糖苷生物合成关键基因的导入和鉴定分析[J]. 生物技术通报, 2023, 39(1): 214-223. |
| [4] | 牛宇辉, 李向茸, 吴贝, 李洪珊, 李殿玉, 陈磊, 魏锁成, 冯若飞. 葡萄糖和丁酸钠对CHO-rHSA工程细胞株中rHSA产量的影响[J]. 生物技术通报, 2022, 38(7): 278-286. |
| [5] | 张国宁, 冯婧娴, 杨颖博, 陈万生, 肖莹. 环糊精葡萄糖基转移酶在天然产物糖基化修饰中的应用[J]. 生物技术通报, 2022, 38(3): 246-255. |
| [6] | 郭宇飞, 闫荣媚, 张小茹, 曹威, 刘浩. 代谢工程改造黑曲霉生产葡萄糖二酸[J]. 生物技术通报, 2022, 38(11): 227-237. |
| [7] | 廖兆民, 蔡俊, 林建国, 杜馨, 王常高. 黑曲霉葡萄糖氧化酶基因在毕赤酵母中的表达及产酶条件的优化[J]. 生物技术通报, 2021, 37(6): 97-107. |
| [8] | 霍明月, 张鸽, 苏玉龙, 李兴, 张海波, 峥嵘, 刘好宝. 一株耐受高浓度葡萄糖且产乙偶姻菌株的筛选及产物鉴定[J]. 生物技术通报, 2020, 36(8): 53-60. |
| [9] | 陈桥, 吴海英, 王宗寿, 谢雨康, 李宜青, 孙俊松. 聚羟基丁酸酯合成引发的高密度生长大肠杆菌的多位点突变分析[J]. 生物技术通报, 2020, 36(7): 112-118. |
| [10] | 董聪, 高庆华, 王玥, 罗同阳. 基于密码子优化的FAD依赖葡萄糖脱氢酶在毕赤酵母中的高效表达及酶学性质[J]. 生物技术通报, 2019, 35(7): 114-120. |
| [11] | 李骥璇, 余磊, 李京美, 郑桂兰, 王洪钟. S-亚胺还原酶和葡萄糖脱氢酶共表达系统的构建及手性胺的合成[J]. 生物技术通报, 2019, 35(1): 105-111. |
| [12] | 高庆华, 董聪, 王玥, 胡美荣, 王庆庆, 王云鹏, 罗同阳, 刘蕾. 共表达分子伴侣PDI和Ero1对葡萄糖氧化酶在毕赤酵母中表达的影响[J]. 生物技术通报, 2018, 34(7): 174-179. |
| [13] | 李真真, 赵佳晨, 马昱澍. 基于sRNA的氧化葡萄糖酸杆菌基因调控的研究[J]. 生物技术通报, 2018, 34(3): 59-66. |
| [14] | 戴晨霞, 曹慧方, 罗会颖, 姚斌, 柏映国, 徐波. 来源于Alicyclobacillus sp.A4葡萄糖异构酶的克隆表达及转化应用研究[J]. 生物技术通报, 2018, 34(11): 144-151. |
| [15] | 冯薇, 胡小妍, 马明娜, 郭萌, 路福平, 李玉. 产β-葡萄糖苷酶细菌的筛选及转化白藜芦醇的研究[J]. 生物技术通报, 2017, 33(11): 130-135. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||