








生物技术通报 ›› 2022, Vol. 38 ›› Issue (1): 205-214.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0137
张廷焕( ), 郭宗义, 柴捷, 潘红梅, 张亮, 陈磊, 龙熙(
), 郭宗义, 柴捷, 潘红梅, 张亮, 陈磊, 龙熙( )
)
收稿日期:2021-02-02
出版日期:2022-01-26
发布日期:2022-02-22
作者简介:张廷焕,男,助理研究员,研究方向:猪功能基因组;E-mail: 基金资助:
ZHANG Ting-huan( ), GUO Zong-yi, CHAI Jie, PAN Hong-mei, ZHANG Liang, CHEN Lei, LONG Xi(
), GUO Zong-yi, CHAI Jie, PAN Hong-mei, ZHANG Liang, CHEN Lei, LONG Xi( )
)
Received:2021-02-02
Published:2022-01-26
Online:2022-02-22
摘要:
本研究旨在探索序列变异对miR-378的结构、表达水平以及靶标关系的影响。利用PCR测序比对不同猪种miR-378的序列突变,预测突变对miR-378二级结构和自由能的改变;构建miR-378表达载体检测突变对其加工过程中各级产物表达水平的作用;运用TargetScan和TargetRank对突变引起的靶标关系变化进行分析;采用microRNA pull-down 技术验证突变对靶标关系的影响。结果发现,在荣昌猪和内江猪群体内pre-miR-378的+49和+68位均发生A>G的序列变异,且改变了pre-miR-378的二级结构和自由能;这两个位点突变后影响了pri-miR-378到pre-miR-378的加工过程,提升了pre-miR-378和成熟体miR-378的表达水平;同时+49/A>G的突变改变了miR-378的功能及其靶标关系,其中GDF6和 RAB10分别为突变后获得和缺失的靶基因,而Runx1t1、Galnt3为不受突变影响的靶基因。本研究的结果强调了miRNAs序列变异的重要性及其对miRNAs生物发生和功能的显著影响。
张廷焕, 郭宗义, 柴捷, 潘红梅, 张亮, 陈磊, 龙熙. 序列变异对miR-378生物发生以及靶标关系的影响[J]. 生物技术通报, 2022, 38(1): 205-214.
ZHANG Ting-huan, GUO Zong-yi, CHAI Jie, PAN Hong-mei, ZHANG Liang, CHEN Lei, LONG Xi. Effects of Sequence Variation on the Biogenesis and Target Relationship of miR-378[J]. Biotechnology Bulletin, 2022, 38(1): 205-214.
| 名称Name | 序列Sequence | 长度 Length/bp |
|---|---|---|
| pri-miR-378-F | CGGGATCCAAGGTATTTGGGGCACTGCA | 23 |
| pri-miR-378-R | GGGAAGCTTCAGGACAGTTCAGGCAAGGT | 20 |
表1 pri-miR-378 PCR扩增引物
Table 1 PCR primers of pri-miR-378
| 名称Name | 序列Sequence | 长度 Length/bp |
|---|---|---|
| pri-miR-378-F | CGGGATCCAAGGTATTTGGGGCACTGCA | 23 |
| pri-miR-378-R | GGGAAGCTTCAGGACAGTTCAGGCAAGGT | 20 |
| 名称Name | 序列Sequence | 长度Length/bp |
|---|---|---|
| pri-miR-378-F | GAGGGAGGCAGCATGGTAAG | 20 |
| pre-miR-378-F | TGAAGGCAGGCAGAACCATT | 20 |
| precursor-miR-378-R | CGGGCCTTCTGACTCCAAG | 19 |
| miR-378/AA | ACTGGACTTGGAGTCAGAAGGC | 22 |
| miR-378/GG | ACTGGGCTTGGAGTCAGAAGGC | 22 |
表2 miR-378 RT-PCR引物
Table 2 RT-PCR primers of miR-378
| 名称Name | 序列Sequence | 长度Length/bp |
|---|---|---|
| pri-miR-378-F | GAGGGAGGCAGCATGGTAAG | 20 |
| pre-miR-378-F | TGAAGGCAGGCAGAACCATT | 20 |
| precursor-miR-378-R | CGGGCCTTCTGACTCCAAG | 19 |
| miR-378/AA | ACTGGACTTGGAGTCAGAAGGC | 22 |
| miR-378/GG | ACTGGGCTTGGAGTCAGAAGGC | 22 |
| 名称Name | 序列Sequence | 长度Length/bp |
|---|---|---|
| GDF6-F | CTCGAGCTACTAAATGACAG | 20 |
| GDF6-R | TCTCCTTCCTCACTGCCTGT | 20 |
| Runx1t1-F | ATCGGGAATTCCTTCACAGGC | 21 |
| Runx1t1-R | GCTTTTTGCAGCTCCGTCAT | 20 |
| Galnt3-F | ACGCAGGTGATTGCTCGTAA | 20 |
| Galnt3-R | AGGTCTGGCACATACGCTTC | 20 |
| RAB10-F | ATGTACTTGCTCAGCTCAACT | 21 |
| RAB10-R | AGGGACTCAAGCACATTATCCA | 22 |
表3 miR-378靶基因RT-PCR扩增引物
Table 3 RT-PCR primers of miR-378 targets
| 名称Name | 序列Sequence | 长度Length/bp |
|---|---|---|
| GDF6-F | CTCGAGCTACTAAATGACAG | 20 |
| GDF6-R | TCTCCTTCCTCACTGCCTGT | 20 |
| Runx1t1-F | ATCGGGAATTCCTTCACAGGC | 21 |
| Runx1t1-R | GCTTTTTGCAGCTCCGTCAT | 20 |
| Galnt3-F | ACGCAGGTGATTGCTCGTAA | 20 |
| Galnt3-R | AGGTCTGGCACATACGCTTC | 20 |
| RAB10-F | ATGTACTTGCTCAGCTCAACT | 21 |
| RAB10-R | AGGGACTCAAGCACATTATCCA | 22 |
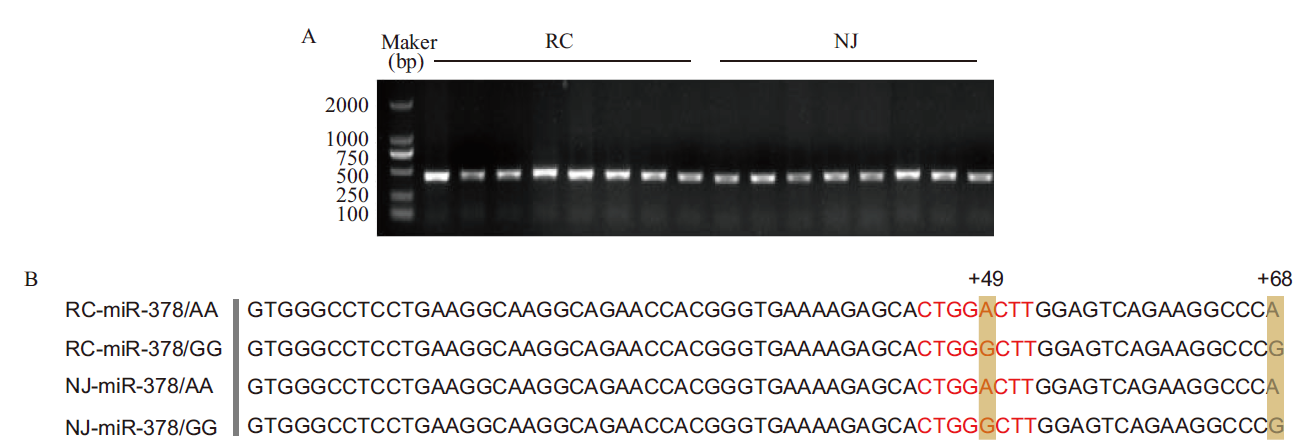
图1 猪miR-378的扩增和测序 A:miR-378 PCR产物的凝胶电泳图;B:不同猪种pre-miR-378序列对比图,红色字母代表miR-378的种子序列。其中,RC代表荣昌猪,NJ代表内江猪,下同
Fig.1 Amplification and sequencing of porcine miR-378 A: Gel electrophoresis of miR-378 PCR products. B: Sequence comparison of pre-miR-378 in different pig breeds and the red letters refer to the seed sequence of miR-378. RC refers to Rongchang pig and NJ refers to Neijiang pig,the same below

图2 miR-378结构和能量预测 A:miR-378的二级结构;B:miR-378的自由能分布。其中,红色方框代表变化最明显的区域
Fig.2 Structure and energy prediction of miR-378 A: Secondary structure of miR-378. B: Energy distribution of miR-378. The red box refers to the areas with the most obvious changes

图3 miR-378过表达载体的构建 A:pri-miR-378 PCR产物的凝胶电泳图;B:过表达载体示意图;C:过表达载体插入片段部分序列图,红色方框代表变异位点
Fig.3 Construction of miR-378 overexpression vector A: Gel electrophoresis of pri-miR-378 PCR products. B: Schematic diagram of overexpression vectors. C: Partial sequence diagram of inserted fragment of overexpression vectors,and the red boxes refer to the mutation sites
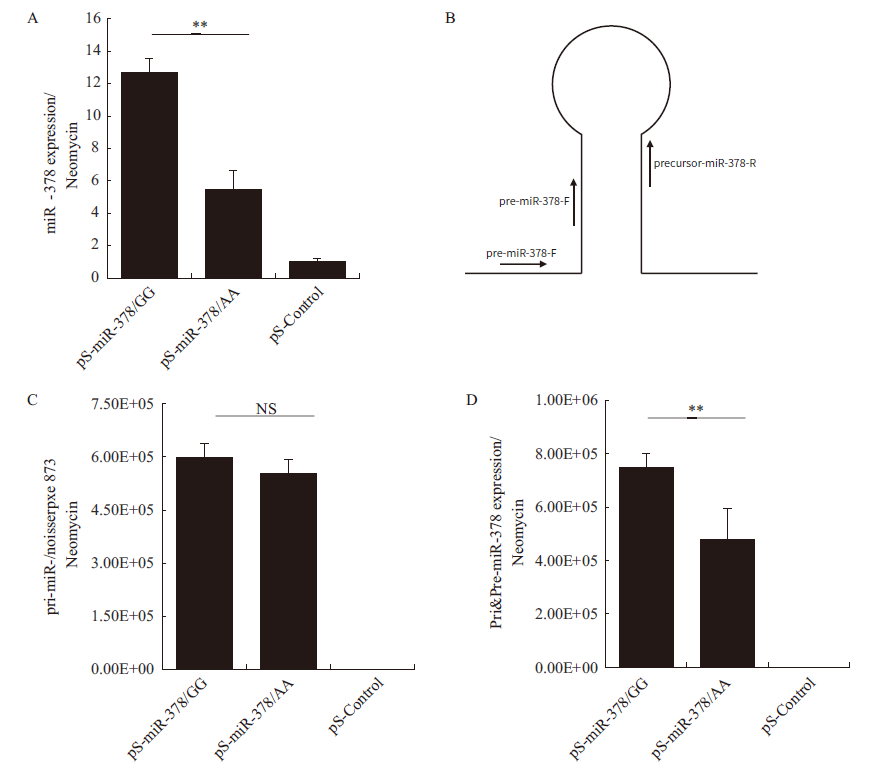
图4 miR-378各级产物表达量分析 A:成熟体miR-378的表达量;B:pri-miR-378和pre-miR-378特异性引物的示意图;C:pri-miR-378的表达量;D:pri-miR-378和pre-miR-378表达总量。其中**代表P<0.01,NS代表P>0.05,下同
Fig.4 Analysis of miR-378 products in expressions A: The expression of mature miR-378. B: Schematic diagram of the specific primers designed to amplify pri-miR-378 and pre-miR-378. C: The expression of pri-miR-378. D: The total expression of pre-miR-378 and pri-miR-378. And ** refers to P < 0.01,NS refers to P > 0.05,the same below
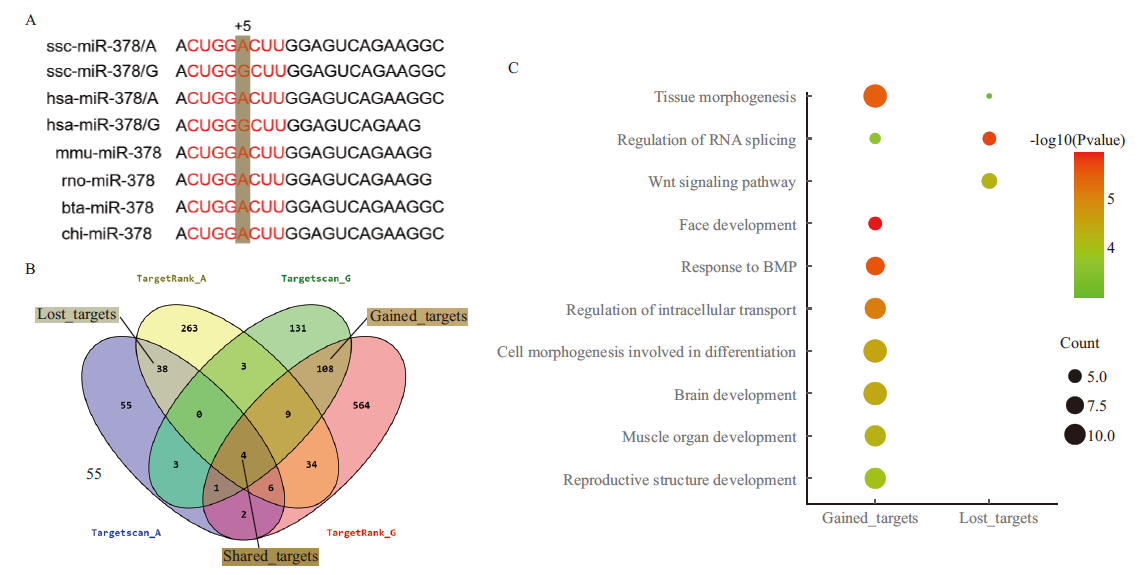
图5 miR-378靶标关系和功能预测 A:不同物种间成熟体miR-378序列比对,红色字母代表种子序列;B:靶基因预测以及不同软件之间的多重比对;C:靶基因的功能富集分析
Fig.5 Target relationship and function prediction of miR-378 A: The sequence alignment of mature miR-378 in different species and the red letters refer to the seed sequence. B: Target gene prediction and multiple comparisons among different softwares. C: Functional enrichment analysis of the targets
| 分组名称 Group name | 靶基因 Target genes |
|---|---|
| 获得的靶基因 Gained-targets | NFIX、ABAT、ADAM19、ASXL3、ATP1B2、AXUD1、BCL2、BMF、BRUNOL6、C14orf83、C15orf27、C9orf41、CACNA1I、CDH1、CHD5、CHRM1、CLCN6、CRISPLD2、DNAJB12、ENG、ETV1、FAIM2、FBXL11、FBXO33、FNDC3A、FRAS1、GUCY1A2、HEYL、IFI30、INPP5D、ITGA5、KCNA5、KIAA0556、LARP1、MGLL、MMP24、MYLK2、NAV1、NCAM1、NOTCH2、NUAK1、OXSR1、PAF1、PAQR4、PDE11A、PELI3、PLEKHA6、PRNPIP、RBM19、REEP2、RHBG、RORC、SFRS7、SHROOM4、SLAIN2、SLC23A2、SNAP29、SOBP、SP1、SPRN、SYT9、TMC1、TMPRSS4、TPI1、TYRO3、UBE2N、WDR78、ZHX3、ZNF609、BSDC1、COBL、CYP26B1、DDX6、ELAVL3、FLJ14154、GDF6、HECTD3、KCNIP3、KIAA0652、LEF1、MAF、MAPK3、PARD6B、PJA2、SBK1、SMARCD2、SP6、TMEM104、TSC22D3、ZNF292、ARNT2、C12orf4、C9orf105、FBXO41、GAB2、GRHL2、KCTD15、KIAA1853、LRRTM3、MTMR3、PAX2、PHACTR2、POLR2J3、RIMS3、SRRM1、THRB、UNC119B、ZCCHC3 |
| 失去的靶基因 Lost-targets | KIAA1522、SDAD1、METTL4、PHC3、SULF1、RBMS3、RRP1B、BMP2、VANGL1、PAPD5、MED12L、PLEKHG2、CBL、GRSF1、SLC38A1、GOLT1A、PLAGL2、SOX7、DCX、DACT1、SLC39A9、KIAA1219、DYRK1A、IPO9、KIAA1576、QSER1、NEK4、TLK2、LBX2、C1orf21、FRMPD4、SFRS3、CALN1、WDR37、DBT、MEF2D、TSPAN17、RAB10 |
| 共有的靶基因 Shared-targets | Runx1t1、Galnt3、KPNA6、BCL2L2 |
表4 miR-378的靶基因列表
Table 4 Target list of miR-378
| 分组名称 Group name | 靶基因 Target genes |
|---|---|
| 获得的靶基因 Gained-targets | NFIX、ABAT、ADAM19、ASXL3、ATP1B2、AXUD1、BCL2、BMF、BRUNOL6、C14orf83、C15orf27、C9orf41、CACNA1I、CDH1、CHD5、CHRM1、CLCN6、CRISPLD2、DNAJB12、ENG、ETV1、FAIM2、FBXL11、FBXO33、FNDC3A、FRAS1、GUCY1A2、HEYL、IFI30、INPP5D、ITGA5、KCNA5、KIAA0556、LARP1、MGLL、MMP24、MYLK2、NAV1、NCAM1、NOTCH2、NUAK1、OXSR1、PAF1、PAQR4、PDE11A、PELI3、PLEKHA6、PRNPIP、RBM19、REEP2、RHBG、RORC、SFRS7、SHROOM4、SLAIN2、SLC23A2、SNAP29、SOBP、SP1、SPRN、SYT9、TMC1、TMPRSS4、TPI1、TYRO3、UBE2N、WDR78、ZHX3、ZNF609、BSDC1、COBL、CYP26B1、DDX6、ELAVL3、FLJ14154、GDF6、HECTD3、KCNIP3、KIAA0652、LEF1、MAF、MAPK3、PARD6B、PJA2、SBK1、SMARCD2、SP6、TMEM104、TSC22D3、ZNF292、ARNT2、C12orf4、C9orf105、FBXO41、GAB2、GRHL2、KCTD15、KIAA1853、LRRTM3、MTMR3、PAX2、PHACTR2、POLR2J3、RIMS3、SRRM1、THRB、UNC119B、ZCCHC3 |
| 失去的靶基因 Lost-targets | KIAA1522、SDAD1、METTL4、PHC3、SULF1、RBMS3、RRP1B、BMP2、VANGL1、PAPD5、MED12L、PLEKHG2、CBL、GRSF1、SLC38A1、GOLT1A、PLAGL2、SOX7、DCX、DACT1、SLC39A9、KIAA1219、DYRK1A、IPO9、KIAA1576、QSER1、NEK4、TLK2、LBX2、C1orf21、FRMPD4、SFRS3、CALN1、WDR37、DBT、MEF2D、TSPAN17、RAB10 |
| 共有的靶基因 Shared-targets | Runx1t1、Galnt3、KPNA6、BCL2L2 |
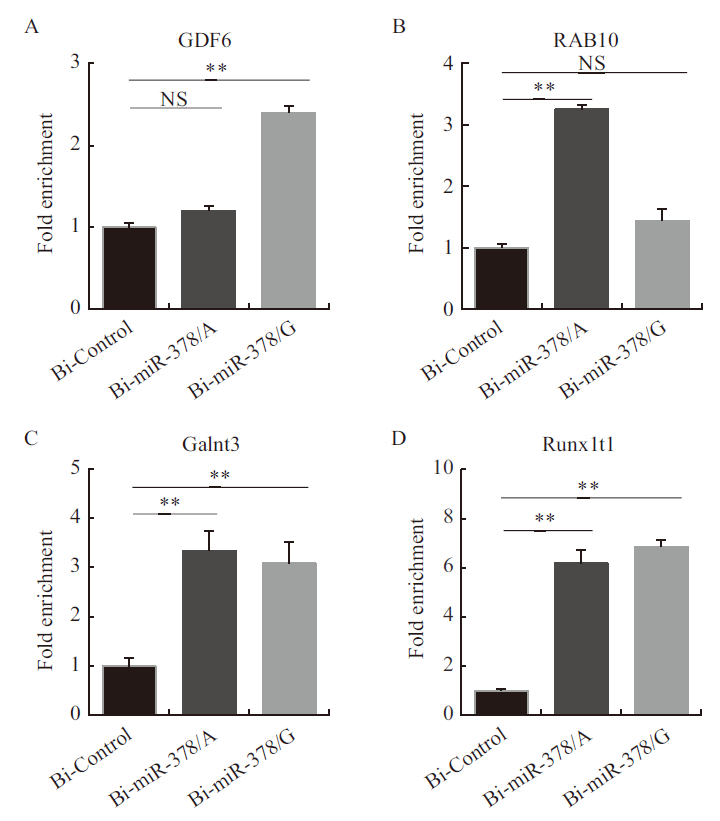
图6 miR-378靶基因的富集程度验证 A:GDF6基因富集程度;B:RAB10基因富集程度;C:Galnt3基因富集程度;D:Runx1t1基因富集程度
Fig.6 Verification of enrichment degree of miR-378 targets A: The enrichment of GDF6 gene. B: The enrichment of RAB10 gene. C: The enrichment of Galnt3 gene. D: The enrichment of Runx1t1 gene
| [1] |
Gebert LFR, MacRae IJ. Regulation of microRNA function in animals[J]. Nat Rev Mol Cell Biol, 2019, 20(1):21-37.
doi: 10.1038/s41580-018-0045-7 URL |
| [2] | Ameres SL, Zamore PD. Diversifying microRNA sequence and function[J]. Nat Rev Mol Cell Biol, 2013, 14(8):475-488. |
| [3] |
Sheng P, Fields C, Aadland K, et al. Dicer cleaves 5'-extended microRNA precursors originating from RNA polymerase II transcription start sites[J]. Nucleic Acids Res, 2018, 46(11):5737-5752.
doi: 10.1093/nar/gky306 URL |
| [4] |
Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs[J]. Nature, 2004, 432(7014):235-240.
doi: 10.1038/nature03120 URL |
| [5] |
Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing[J]. Nature, 2003, 425(6956):415-419.
doi: 10.1038/nature01957 URL |
| [6] |
Fareh M, Yeom KH, Haagsma AC, et al. TRBP ensures efficient Dicer processing of precursor microRNA in RNA-crowded environments[J]. Nat Commun, 2016, 7:13694.
doi: 10.1038/ncomms13694 URL |
| [7] |
Koscianska E, Starega-Roslan J, Krzyzosiak WJ. The role of Dicer protein partners in the processing of microRNA precursors[J]. PLoS One, 2011, 6(12):e28548.
doi: 10.1371/journal.pone.0028548 URL |
| [8] |
Sun G, Yan J, Noltner K, et al. SNPs in human miRNA genes affect biogenesis and function[J]. RNA, 2009, 15(9):1640-1651.
doi: 10.1261/rna.1560209 URL |
| [9] |
Kotani A, Ha D, Schotte D, et al. A novel mutation in the miR-128b gene reduces miRNA processing and leads to glucocorticoid resistance of MLL-AF4 acute lymphocytic leukemia cells[J]. Cell Cycle, 2010, 9(6):1037-1042.
pmid: 20237425 |
| [10] |
Duan RH, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA[J]. Hum Mol Genet, 2007, 16(9):1124-1131.
doi: 10.1093/hmg/ddm062 URL |
| [11] |
Harnprasopwat R, Ha D, Toyoshima T, et al. Alteration of processing induced by a single nucleotide polymorphism in pri-miR-126[J]. Biochem Biophys Res Commun, 2010, 399(2):117-122.
doi: 10.1016/j.bbrc.2010.07.009 URL |
| [12] |
Zhang Y, Yun Z, Gong L, et al. Comparison of miRNA evolution and function in plants and animals[J]. Microrna, 2018, 7(1):4-10.
doi: 10.2174/2211536607666180126163031 pmid: 29372676 |
| [13] |
Mencía Á, Modamio-Høybjør S, Redshaw N, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss[J]. Nat Genet, 2009, 41(5):609-613.
doi: 10.1038/ng.355 URL |
| [14] |
Xu LL, Shi CM, Xu GF, et al. TNF-α, IL-6, and leptin increase the expression of miR-378, an adipogenesis-related microRNA in human adipocytes[J]. Cell Biochem Biophys, 2014, 70(2):771-776.
doi: 10.1007/s12013-014-9980-x URL |
| [15] |
Gerin I, Bommer GT, McCoin CS, et al. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis[J]. Am J Physiol Endocrinol Metab, 2010, 299(2):E198-E206.
doi: 10.1152/ajpendo.00179.2010 URL |
| [16] |
Pan D, Mao C, Quattrochi B, et al. MicroRNA-378 controls classical brown fat expansion to counteract obesity[J]. Nat Commun, 2014, 5:4725.
doi: 10.1038/ncomms5725 URL |
| [17] |
Gagan J, Dey BK, Layer R, et al. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation[J]. J Biol Chem, 2011, 286(22):19431-19438.
doi: 10.1074/jbc.M111.219006 URL |
| [18] |
Hou X, Tang Z, Liu H, et al. Discovery of MicroRNAs associated with myogenesis by deep sequencing of serial developmental skeletal muscles in pigs[J]. PLoS One, 2012, 7(12):e52123.
doi: 10.1371/journal.pone.0052123 URL |
| [19] |
Knezevic I, Patel A, Sundaresan NR, et al. A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor:implications in postnatal cardiac remodeling and cell survival[J]. J Biol Chem, 2012, 287(16):12913-12926.
doi: 10.1074/jbc.M111.331751 pmid: 22367207 |
| [20] |
Liu W, Cao H, Ye C, et al. Hepatic miR-378 targets p110alpha and controls glucose and lipid homeostasis by modulating hepatic insulin signalling[J]. Nat Commun, 2014, 5:5684.
doi: 10.1038/ncomms6684 URL |
| [21] |
Xu S, Linher-Melville K, Yang BB, et al. Micro-RNA378(miR-378)regulates ovarian estradiol production by targeting aromatase[J]. Endocrinology, 2011, 152(10):3941-3951.
doi: 10.1210/en.2011-1147 URL |
| [22] |
Ma T, Jiang H, Gao Y, et al. Microarray analysis of differentially expressed microRNAs in non-regressed and regressed bovine corpus luteum tissue;microRNA-378 may suppress luteal cell apoptosis by targeting the interferon gamma receptor 1 gene[J]. J Appl Genet, 2011, 52(4):481-486.
doi: 10.1007/s13353-011-0055-z URL |
| [23] |
Chai J, Chen L, Luo Z, et al. Spontaneous single nucleotide polymorphism in porcine microRNA-378 seed region leads to functional alteration[J]. Biosci Biotechnol Biochem, 2018, 82(7):1081-1089.
doi: 10.1080/09168451.2018.1459175 URL |
| [24] |
Sun W, Lan J, Chen L, et al. A mutation in porcine pre-miR-15b alters the biogenesis of MiR-15b\16-1 cluster and strand selection of MiR-15b[J]. PLoS One, 2017, 12(5):e0178045.
doi: 10.1371/journal.pone.0178045 URL |
| [25] |
Li Y, Wang XQ, Zhang L, et al. A SNP in pri-miR-10a is associated with recurrent spontaneous abortion in a Han-Chinese population[J]. Oncotarget, 2016, 7(7):8208-8222.
doi: 10.18632/oncotarget.v7i7 URL |
| [26] |
Locke JM, Lango Allen H, Harries LW. A rare SNP in pre-miR-34a is associated with increased levels of miR-34a in pancreatic beta cells[J]. Acta Diabetol, 2014, 51(2):325-329.
doi: 10.1007/s00592-013-0499-1 URL |
| [27] |
Gong J, Tong Y, Zhang HM, et al. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis[J]. Hum Mutat, 2012, 33(1):254-263.
doi: 10.1002/humu.21641 URL |
| [28] |
Hughes AE, Bradley DT, Campbell M, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract[J]. Am J Hum Genet, 2011, 89(5):628-633.
doi: 10.1016/j.ajhg.2011.09.014 pmid: 21996275 |
| [29] |
Dorn GW, Matkovich SJ, Eschenbacher WH, et al. A human 3' miR-499 mutation alters cardiac mRNA targeting and function[J]. Circ Res, 2012, 110(7):958-967.
doi: 10.1161/CIRCRESAHA.111.260752 URL |
| [30] |
Thielen N, van der Kraan P, van Caam A. TGFβ/BMP signaling pathway in cartilage homeostasis[J]. Cells, 2019, 8(9):969.
doi: 10.3390/cells8090969 URL |
| [31] |
Sartori R, Schirwis E, Blaauw B, et al. BMP signaling controls muscle mass[J]. Nat Genet, 2013, 45(11):1309-1318.
doi: 10.1038/ng.2772 URL |
| [32] |
Clendenning DE, Mortlock DP. The BMP ligand Gdf6 prevents differentiation of coronal suture mesenchyme in early cranial development[J]. PLoS One, 2012, 7(5):e36789.
doi: 10.1371/journal.pone.0036789 URL |
| [33] |
Logan CY, Nusse R. The Wnt signaling pathway in development and disease[J]. Annu Rev Cell Dev Biol, 2004, 20:781-810.
doi: 10.1146/cellbio.2004.20.issue-1 URL |
| [34] |
Ghosh N, Hossain U, Mandal A, et al. The Wnt signaling pathway:a potential therapeutic target against cancer[J]. Ann N Y Acad Sci, 2019, 1443(1):54-74.
doi: 10.1111/nyas.2019.1443.issue-1 URL |
| [35] | O’Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation[J]. Front Endocrinol:Lausanne, 2018, 9:402. |
| [36] |
Cai Y, Yu X, Hu S, et al. A brief review on the mechanisms of miRNA regulation[J]. Genomics Proteomics Bioinformatics, 2009, 7(4):147-154.
doi: 10.1016/S1672-0229(08)60044-3 URL |
| [1] | 史建磊, 宰文珊, 苏世闻, 付存念, 熊自立. 番茄青枯病抗性相关miRNA的鉴定与表达分析[J]. 生物技术通报, 2023, 39(5): 233-242. |
| [2] | 王楠楠, 王文佳, 朱强. 植物胁迫相关microRNA研究进展[J]. 生物技术通报, 2022, 38(8): 1-11. |
| [3] | 寇航, 王艳梅, 李彤, 薄明井, 张惟材, 熊向华, 黎明. 基于Methylovorus sp. J1-1基因组尺度代谢网络优化吡咯喹啉醌合成[J]. 生物技术通报, 2022, 38(2): 173-183. |
| [4] | 洪军, 卫夏怡, 吉冰洁, 叶延欣, 程天赐. 铜绿假单胞菌对鲎素耐药前后的差异表达基因及SNP变化研究[J]. 生物技术通报, 2021, 37(9): 191-202. |
| [5] | 张廷焕, 张利娟, 陈四清, 郭宗义. 猪miR-378种子序列的多态性对其功能以及胴体性状的影响[J]. 生物技术通报, 2021, 37(6): 154-162. |
| [6] | 李玲, 杨丽霞, 郭梅. CNR转录因子在番茄果实成熟过程中的功能[J]. 生物技术通报, 2021, 37(2): 51-62. |
| [7] | 张廷焕, 龙熙, 郭宗义, 柴捷. miR-378促进脂质生成相关靶基因鉴定[J]. 生物技术通报, 2021, 37(2): 80-87. |
| [8] | 李泽卿, 刘彩贤, 邢文, 文亚峰. miRNA在植物响应高温胁迫中的研究进展[J]. 生物技术通报, 2020, 36(2): 149-157. |
| [9] | 胡积祥, 曹雅倩, 朱秀梅, 余超, 田芳, 杨凤环, 陈华民, 何晨阳. 基于瞬时表达系统的水稻miRNA靶基因快速验证系统的建立[J]. 生物技术通报, 2019, 35(10): 57-63. |
| [10] | 纪会, 王会, 柴志欣, 王吉坤, 罗晓林, 姬秋梅, 信金伟, 钟金城. 牦牛miR-378前体克隆及组织表达分析[J]. 生物技术通报, 2019, 35(1): 58-66. |
| [11] | 谢洁 ,王明 ,李青 ,潘妃 ,熊兴耀 ,秦玉芝. 植物miR390的研究进展[J]. 生物技术通报, 2018, 34(6): 1-10. |
| [12] | 刘伟灿,周永刚,王兴超,王法微,王南,董园园,李晓薇,李海燕. 植物MicroRNA介导的基因调控在作物改良中的应用潜能[J]. 生物技术通报, 2016, 32(4): 6-15. |
| [13] | 曾长英, 周玉飞, 彭明. miR319a及其靶基因在木薯中的低温响应分析[J]. 生物技术通报, 2015, 31(11): 173-178. |
| [14] | 王维,张玉娟,陈洁,刘聚波,夏民旋,沈法富. 植物逆境胁迫相关miRNA研究进展[J]. 生物技术通报, 2015, 31(1): 1-10. |
| [15] | 李静,黄英,黄丽梅,杨明华,李琦华,贾俊静,赵素梅. miR-143在脂肪细胞分化和脂类代谢中的作用[J]. 生物技术通报, 2014, 0(9): 34-38. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||