








生物技术通报 ›› 2022, Vol. 38 ›› Issue (4): 303-310.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0906
周晓楠1,2( ), 徐金青1,3, 雷雨晴1,2, 王海庆1,3(
), 徐金青1,3, 雷雨晴1,2, 王海庆1,3( )
)
收稿日期:2021-07-15
出版日期:2022-04-26
发布日期:2022-05-06
通讯作者:
王海庆,男,博士,研究员,研究方向:豆科牧草资源评价与抗逆改良;E-mail: wanghq@nwipb.cas.cn作者简介:周晓楠,女,硕士研究生,研究方向:豆科牧草资源利用与评价;E-mail: 1186759459@qq.com
基金资助:
ZHOU Xiao-nan1,2( ), XU Jin-qing1,3, LEI Yu-qing1,2, WANG Hai-qing1,3(
), XU Jin-qing1,3, LEI Yu-qing1,2, WANG Hai-qing1,3( )
)
Received:2021-07-15
Published:2022-04-26
Online:2022-05-06
摘要:
开发青藏扁蓿豆(Medicago archiducis-nicolai)单核苷酸多态性标记(single nucleotide polymorphism,SNP),并开展遗传多样性初步分析,为青藏扁蓿豆种质资源的评价与利用提供理论支持。以5个青藏扁蓿豆群体共80份材料进行GBS(genotyping-by-sequencing)测序,采用GATK软件开发SNP标记,并以此进行遗传结构和遗传多样性的初步分析。测序产生的序列数据量为60.79 Gb,过滤后共获得12 796个SNP位点,且SNP突变类型存在明显的转换型偏差现象。不同地理来源青藏扁蓿豆群体观测杂合度(HO)为0.187 68-0.304 36,期望杂合度(HE)为0.201 97-0.364 34,遗传多样性指数(Pi)为0.178 32-0.241 34;祁连(QLCD)群体具有相对最高的遗传多样性水平。遗传结构分析表明5个群体分为2组,且群体遗传距离与地理距离之间存在极显著的正相关性。SNP标记适用于青藏扁蓿豆遗传多样性分析。
周晓楠, 徐金青, 雷雨晴, 王海庆. 基于GBS-seq的青藏扁蓿豆SNP标记开发[J]. 生物技术通报, 2022, 38(4): 303-310.
ZHOU Xiao-nan, XU Jin-qing, LEI Yu-qing, WANG Hai-qing. Development of SNP Markers in Medicago archiducis-nicolai Based on GBS-seq[J]. Biotechnology Bulletin, 2022, 38(4): 303-310.
| 采样地Location | 经度Latitude(E) | 纬度Longitude(N) | 海拔Altitude/m | 样本量Sample size |
|---|---|---|---|---|
| 青海日月山RYS | 101°7'11.7″ | 36°26'50.6″ | 3 311 | 16 |
| 青海祁连QLCD | 100°28'3″ | 38°03'40″ | 3 008 | 16 |
| 青海大通DTXR | 101°24'24″ | 37°1'1.7″ | 2 954 | 16 |
| 青海湟源HY | 101°13'9″ | 36°34'30.3″ | 2 841 | 16 |
| 青海西宁西山XNXS | 101°44'17.5″ | 36°37'17.5″ | 2 462 | 16 |
表1 青藏扁蓿豆材料来源
Table 1 Sources of M. archiducis-nicolai materials
| 采样地Location | 经度Latitude(E) | 纬度Longitude(N) | 海拔Altitude/m | 样本量Sample size |
|---|---|---|---|---|
| 青海日月山RYS | 101°7'11.7″ | 36°26'50.6″ | 3 311 | 16 |
| 青海祁连QLCD | 100°28'3″ | 38°03'40″ | 3 008 | 16 |
| 青海大通DTXR | 101°24'24″ | 37°1'1.7″ | 2 954 | 16 |
| 青海湟源HY | 101°13'9″ | 36°34'30.3″ | 2 841 | 16 |
| 青海西宁西山XNXS | 101°44'17.5″ | 36°37'17.5″ | 2 462 | 16 |
| 群体名称 Population name | 测序片段数 Raw reads | 碱基数 Raw bases/Gb | 过滤后测序片段数Clean reads | 过滤后碱基数Clean bases/Gb | 酶捕获率 Enzyme catch ratio/% | 测序质量 Sequencing quality Q30/% | GC含量 GC content/% |
|---|---|---|---|---|---|---|---|
| RYS | 4 815 069 | 0.72 | 3 856 582 | 0.58 | 99.39 | 93.80 | 53.23 |
| XNXS | 4 360 916 | 0.65 | 3 468 857 | 0.52 | 98.74 | 93.13 | 46.40 |
| HY | 6 638 025 | 0.99 | 5 456 407 | 0.82 | 98.85 | 93.39 | 49.90 |
| QLCD | 4 800 074 | 0.71 | 3 981 534 | 0.60 | 99.16 | 93.07 | 42.29 |
| DTXR | 4 884 748 | 0.73 | 4 044 807 | 0.61 | 99.46 | 93.33 | 51.73 |
表2 青藏扁蓿豆群体测序数据统计
Table 2 Summary of sequencing data of M. archiducis-nicolai populations
| 群体名称 Population name | 测序片段数 Raw reads | 碱基数 Raw bases/Gb | 过滤后测序片段数Clean reads | 过滤后碱基数Clean bases/Gb | 酶捕获率 Enzyme catch ratio/% | 测序质量 Sequencing quality Q30/% | GC含量 GC content/% |
|---|---|---|---|---|---|---|---|
| RYS | 4 815 069 | 0.72 | 3 856 582 | 0.58 | 99.39 | 93.80 | 53.23 |
| XNXS | 4 360 916 | 0.65 | 3 468 857 | 0.52 | 98.74 | 93.13 | 46.40 |
| HY | 6 638 025 | 0.99 | 5 456 407 | 0.82 | 98.85 | 93.39 | 49.90 |
| QLCD | 4 800 074 | 0.71 | 3 981 534 | 0.60 | 99.16 | 93.07 | 42.29 |
| DTXR | 4 884 748 | 0.73 | 4 044 807 | 0.61 | 99.46 | 93.33 | 51.73 |
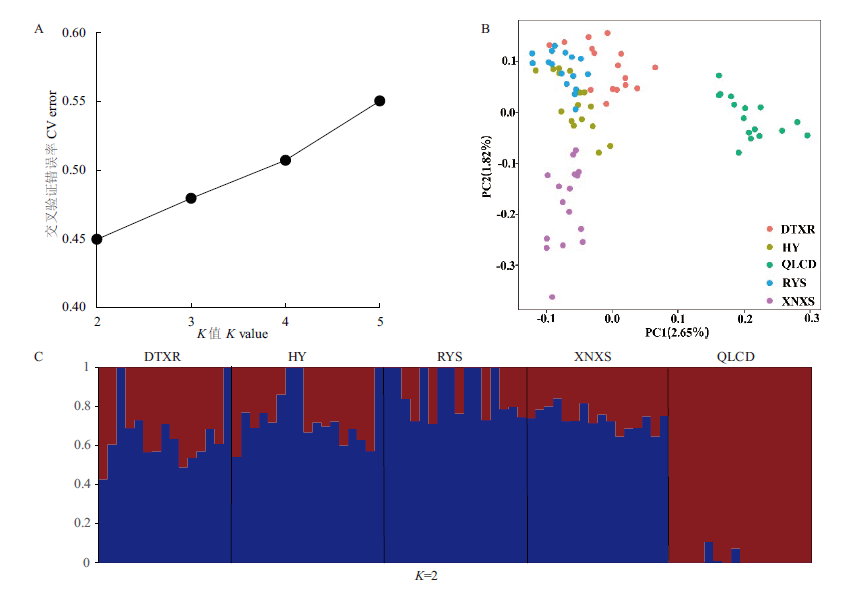
图2 群体遗传结构分析 A:利用ADMIXTURE软件对80份青藏扁蓿豆材料进行群体结构分析,计算K为2-5时的CV error;B:对80份青藏扁蓿豆进行主成分分析;C:K=2时80份青藏扁蓿豆的群体结构
Fig.2 Population genetic structure of populations A:ADMIXTURE software was used to analyze the population structure of 80 M.archiducis-nicolai materials,and the cross-validation(CV)error was calculated when K was 2-5. B:Principal component analysis(PCA)of the 80 M.archiducis-nicolai materials. C:The population structure of 80 M. archiducis-nicolai at K=2
| 群体 Population | HY | RYS | QLCD | XNXS | DTXR |
|---|---|---|---|---|---|
| HY | 0.0000 | 0.0093** | 0.0282** | 0.0138** | 0.0146** |
| RYS | 0.0000 | 0.0369** | 0.0213** | 0.0171** | |
| QLCD | 0.0000 | 0.0338** | 0.0258** | ||
| XNXS | 0.0000 | 0.0215** | |||
| DTXR | 0.0000 |
表3 群体间遗传分化指数(FST)
Table 3 Index of genetic differentiation(FST)between populations
| 群体 Population | HY | RYS | QLCD | XNXS | DTXR |
|---|---|---|---|---|---|
| HY | 0.0000 | 0.0093** | 0.0282** | 0.0138** | 0.0146** |
| RYS | 0.0000 | 0.0369** | 0.0213** | 0.0171** | |
| QLCD | 0.0000 | 0.0338** | 0.0258** | ||
| XNXS | 0.0000 | 0.0215** | |||
| DTXR | 0.0000 |
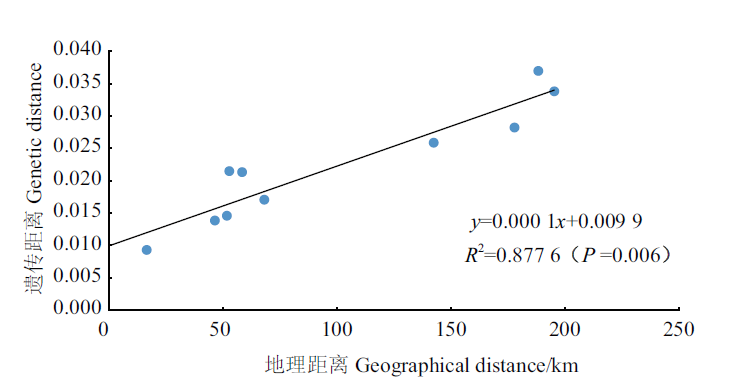
图3 群体遗传距离与地理距离的相关性 注:**:群体间存在极显著(P<0.01)遗传分化
Fig.3 Correlation between population genetic distance and geographical distance Note:** indicate extremely significant differentiation among populations at 0.01 levels,respectively
| 群体 Population | 等位基因数 Number of alleles(NA) | 观测杂合度 Observed heterozygosity(HO) | 期望杂合度 Expected heterozygosity(HE) | 遗传多样性指数 Nucleotide diversity index(Pi) |
|---|---|---|---|---|
| DTXR | 2 | 0.224 02 | 0.222 41 | 0.202 46 |
| QLCD | 2 | 0.304 36 | 0.364 34 | 0.241 34 |
| XNXS | 2 | 0.243 44 | 0.234 05 | 0.213 86 |
| HY | 2 | 0.234 03 | 0.226 40 | 0.209 63 |
| RYS | 2 | 0.187 68 | 0.201 97 | 0.178 32 |
表4 青藏扁蓿豆野生群体遗传参数统计
Table 4 Statistics of genetic parameters in M. archiducis-nicolai wild populations
| 群体 Population | 等位基因数 Number of alleles(NA) | 观测杂合度 Observed heterozygosity(HO) | 期望杂合度 Expected heterozygosity(HE) | 遗传多样性指数 Nucleotide diversity index(Pi) |
|---|---|---|---|---|
| DTXR | 2 | 0.224 02 | 0.222 41 | 0.202 46 |
| QLCD | 2 | 0.304 36 | 0.364 34 | 0.241 34 |
| XNXS | 2 | 0.243 44 | 0.234 05 | 0.213 86 |
| HY | 2 | 0.234 03 | 0.226 40 | 0.209 63 |
| RYS | 2 | 0.187 68 | 0.201 97 | 0.178 32 |
| [1] |
Small E, Jomphe M. A synopsis of the genus Medicago(Leguminosae)[J]. Can J Bot, 1989, 67(11):3260-3294.
doi: 10.1139/b89-405 URL |
| [2] |
Jin DM, Ma JJ, Ma WH, et al. Legumes in Chinese natural grasslands:species, biomass, and distribution[J]. Rangel Ecol Manag, 2013, 66(6):648-656.
doi: 10.2111/REM-D-12-00159.1 URL |
| [3] |
Wu XP, Liu DM, Gulzar K, et al. Population genetic structure and demographic history of Medicago ruthenica(Fabaceae)on the Qinghai-Tibetan Plateau based on nuclear ITS and chloroplast markers[J]. Biochem Syst Ecol, 2016, 69:204-212.
doi: 10.1016/j.bse.2016.10.005 URL |
| [4] |
Robledo-Arnuncio JJ, Klein EK, Muller-Landau HC, et al. Space, time and complexity in plant dispersal ecology[J]. Mov Ecol, 2014, 2(1):16.
doi: 10.1186/s40462-014-0016-3 pmid: 25709828 |
| [5] | 吴小培, 沈迎芳, 王海庆. 基于trnL-trnF序列的扁蓿豆和青藏扁蓿豆遗传多样性及其群体遗传结构分析[J]. 草业科学, 2016, 33(6):1136-1146. |
| Wu XP, Shen YF, Wang HQ. Analysis of genetic diversity and population genetic structure of Medicago archiducis-nolai and Medicago ruthenica populations based on cpDNA trnL-trnF sequences[J]. Pratacultural Sci, 2016, 33(6):1136-1146. | |
| [6] | 王英芳, 张业猛, 刘德梅, 等. 基于转录组测序的青藏扁蓿豆EST-SSR标记开发与验证[J]. 草业科学, 2020, 37(4):718-727. |
| Wang YF, Zhang YM, Liu DM, et al. Development and verification of EST-SSR markers in Medicago archiducis-nicolai by transcriptome sequencing[J]. Pratacultural Sci, 2020, 37(4):718-727. | |
| [7] |
Wang YF, Shen YF, Liu DM, et al. Insights from putatively neutral EST-SSR markers on the population genetic structure and genetic diversity of the Qinghai-Tibetan Plateau endemic Medicago archiducis-nicolai Sirjaev[J]. Genet Resour Crop Evol, 2021, 68(6):2537-2548.
doi: 10.1007/s10722-021-01147-y URL |
| [8] |
Barendse W, Harrison BE, Bunch RJ, et al. Genome wide signatures of positive selection:the comparison of independent samples and the identification of regions associated to traits[J]. BMC Genomics, 2009, 10:178.
doi: 10.1186/1471-2164-10-178 pmid: 19393047 |
| [9] |
Singh A, Singh PK, Singh R, et al. SNP haplotypes of the BADH1 gene and their association with aroma in rice(Oryza sativa L.)[J]. Mol Breed, 2010, 26(2):325-338.
doi: 10.1007/s11032-010-9425-1 URL |
| [10] |
Stephens JC, Schneider JA, Tanguay DA, et al. Haplotype variation and linkage disequilibrium in 313 human genes[J]. Science, 2001, 293(5529):489-493.
doi: 10.1126/science.1059431 pmid: 11452081 |
| [11] |
Hyten DL, Choi IY, Song QJ, et al. A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus mapping[J]. Crop Sci, 2010, 50(3):960-968.
doi: 10.2135/cropsci2009.06.0360 URL |
| [12] | 申淑慧, 戴习林. 基于生长和抗逆功能基因SNP分子标记的凡纳滨对虾野生及选育群体遗传多样性分析[J]. 南方农业学报, 2020, 51(11):2836-2845. |
| Shen SH, Dai XL. Genetic diversity analysis in wild and selected populations of Penaeus vannamei based on SNP markers in growth and stress resistance genes[J]. J South Agric, 2020, 51(11):2836-2845. | |
| [13] | Glaubitz JC, Casstevens TM, Lu F, et al. TASSEL-GBS:a high capacity genotyping by sequencing analysis pipeline[J]. PLoS One, 2014, 9(2):e90346. |
| [14] | Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue[J]. Phytochemical Bulletin, 1987, 19:11-15. |
| [15] | Elshire RJ, Glaubitz JC, Sun Q, et al. A robust, simple genotyping-by-sequencing(GBS)approach for high diversity species[J]. PLoS One, 2011, 6(5):e19379. |
| [16] |
Catchen J, Hohenlohe PA, Bascham S, et al. Stacks:an analysis tool set for population genomics[J]. Mol Ecol, 2013, 22(11):3124-3140.
doi: 10.1111/mec.12354 pmid: 23701397 |
| [17] | Ginestet C. ggplot2:elegant graphics for data analysis[J]. J Royal Stat Soc:Ser A Stat Soc, 2011, 174(1):245-246. |
| [18] |
Excoffier L, Lischer HE. Arlequin suite ver 3. 5:a new series of programs to perform population genetics analyses under Linux and Windows[J]. Mol Ecol Resour, 2010, 10(3):564-567.
doi: 10.1111/j.1755-0998.2010.02847.x pmid: 21565059 |
| [19] |
Peakall R, Smouse PE. GenAlEx 6. 5:genetic analysis in Excel. Population genetic software for teaching and research——an update[J]. Bioinformatics, 2012, 28(19):2537-2539.
pmid: 22820204 |
| [20] | 蔡露, 杨欢, 王勇, 等. 利用GBS技术开发烟草SNP标记及遗传多样性分析[J]. 中国烟草科学, 2018, 39(5):17-24. |
| Cai L, Yang H, Wang Y, et al. Analysis of genetic diversity of tobacco germplasm resources based on SNP markers via genotyping-by-sequencing technology[J]. Chin Tob Sci, 2018, 39(5):17-24. | |
| [21] |
Yu LX, Zheng P, Zhang T, et al. Genotyping-by-sequencing-based genome-wide association studies on Verticillium wilt resistance in autotetraploid alfalfa(Medicago sativa L.)[J]. Mol Plant Pathol, 2017, 18(2):187-194.
doi: 10.1111/mpp.12389 URL |
| [22] |
Zhao H, Li QZ, Li J, et al. The study of neighboring nucleotide composition and transition/transversion bias[J]. Sci China Ser C:Life Sci, 2006, 49(4):395-402.
doi: 10.1007/s11427-006-2002-5 URL |
| [23] | 严佳文, 肖图舰, 袁启凤, 等. 基于转录组序列的火龙果SSR和SNP多态性分析[J]. 热带作物学报, 2018, 39(7):1338-1343. |
| Yan JW, Xiao TJ, Yuan QF, et al. SSR and SNP polymorphism analysis based on Hylocereus polyrhizus transcriptome[J]. Chin J Trop Crops, 2018, 39(7):1338-1343. | |
| [24] |
Collins DW, Jukes TH. Rates of transition and transversion in coding sequences since the human-rodent divergence[J]. Genomics, 1994, 20(3):386-396.
pmid: 8034311 |
| [25] |
Gojobori T, Li WH, Graur D. Patterns of nucleotide substitution in Pseudogenes and functional genes[J]. J Mol Evol, 1982, 18(5):360-369.
pmid: 7120431 |
| [26] |
Li WH, Wu CI, Luo CC. Nonrandomness of point mutation as reflected in nucleotide substitutions in Pseudogenes and its evolutionary implications[J]. J Mol Evol, 1984, 21(1):58-71.
pmid: 6442359 |
| [27] |
Zhao Z, Boerwinkle E. Neighboring-nucleotide effects on single nucleotide polymorphisms:a study of 2. 6 million polymorphisms across the human genome[J]. Genome Res, 2002, 12(11):1679-1686.
doi: 10.1101/gr.287302 URL |
| [28] |
Stojak J, Borowik T, Górny M, et al. Climatic influences on the genetic structure and distribution of the common vole and field vole in Europe[J]. Mammal Res, 2019, 64(1):19-29.
doi: 10.1007/s13364-018-0395-8 URL |
| [29] |
McCauley DE. What is the influence of the seed bank on the persistence and genetic structure of plant populations that experience a high level of disturbance?[J]. New Phytol, 2014, 202(3):734-735.
doi: 10.1111/nph.12732 pmid: 24716514 |
| [30] |
Hamrick JL, Trapnell DW. Using population genetic analyses to understand seed dispersal patterns[J]. Acta Oecologica, 2011, 37(6):641-649.
doi: 10.1016/j.actao.2011.05.008 URL |
| [31] |
Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance[J]. Genetics, 1997, 145(4):1219-1228.
doi: 10.1093/genetics/145.4.1219 pmid: 9093870 |
| [32] | Wright S. Variability within and among natural populations[M]. Chicago: University of Chicago Press, 1978. |
| [33] | Favre A, Päckert M, Pauls SU, et al. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas[J]. Biol Rev, 2015, 90(1):236-253. |
| [1] | 崔学强, 黄昌艳, 邓杰玲, 李先民, 李秀玲, 张自斌. 基于SLAF-seq技术的石斛兰SNP标记开发及亲缘关系分析[J]. 生物技术通报, 2023, 39(6): 141-148. |
| [2] | 肖小军, 陈明, 韩德鹏, 余跑兰, 郑伟, 肖国滨, 周庆红, 周会汶. 甘蓝型油菜每角果粒数全基因组关联分析[J]. 生物技术通报, 2023, 39(3): 143-151. |
| [3] | 洪军, 卫夏怡, 吉冰洁, 叶延欣, 程天赐. 铜绿假单胞菌对鲎素耐药前后的差异表达基因及SNP变化研究[J]. 生物技术通报, 2021, 37(9): 191-202. |
| [4] | 王衍莉, 杨义明, 范书田, 赵滢, 许培磊, 路文鹏, 李昌禹. 基于SSR分子标记的73份山葡萄及杂交后代的遗传多样性分析[J]. 生物技术通报, 2021, 37(1): 189-197. |
| [5] | 陈一丹, 张昱, 杨洁, 张勤, 姜力. 基于转录组测序的奶牛产奶性状重要功能基因挖掘[J]. 生物技术通报, 2020, 36(9): 244-252. |
| [6] | 张乐超, 刘月琴, 段春辉, 张英杰, 王泳, 郭云霞. 7个地方山羊品种遗传多样性及遗传结构分析[J]. 生物技术通报, 2020, 36(6): 183-190. |
| [7] | 余钧剑, 迟美丽, 贾永义, 刘士力, 竺俊全, 顾志敏. 四引物扩增受阻突变体系PCR技术及其在动植物遗传育种研究中的应用[J]. 生物技术通报, 2020, 36(5): 32-38. |
| [8] | 李勇慧, 于相丽, 马会萍, 高凯, 刘名雪. 不同品种牡丹ISSR遗传多样性分析[J]. 生物技术通报, 2020, 36(4): 78-83. |
| [9] | 张一中, 范昕琦, 杨慧勇, 张晓娟, 邵强, 梁笃, 郭琦, 柳青山, 杜维俊. 基于简化基因组测序高粱育种材料亲缘关系的分析[J]. 生物技术通报, 2020, 36(12): 21-33. |
| [10] | 邹坤, 崔红艳, 薛缘, 张少伟, 路丽丽, 赵志辉, 苏瑛. 雷州黑鸭FSHR和ESR1与繁殖性状关联分析[J]. 生物技术通报, 2019, 35(8): 118-126. |
| [11] | 赵昕鹏, 周云, 吕琳琳, 李锁平, 张大乐. 节节麦遗传多样性及在改良普通小麦中的应用[J]. 生物技术通报, 2019, 35(7): 181-189. |
| [12] | 郭萌萌, 周延清, 段红英, 杨珂, 邵露营. 基于地黄转录组数据的SNP标记开发与地黄指纹图谱构建[J]. 生物技术通报, 2019, 35(11): 224-230. |
| [13] | 石建斌, 周红, 王宁, 许庆华, 乔文青, 严根土. 棉花SSR标记种质资源纯度鉴定及遗传多样性分析[J]. 生物技术通报, 2018, 34(7): 138-146. |
| [14] | 赵宁, 陈双林. 竹黄遗传多样性与竹红菌素合成和抗癌作用的研究进展[J]. 生物技术通报, 2018, 34(4): 16-23. |
| [15] | 武海月, 赵爽, 刘宇, 李华敏, 宋爽, 牛玉蓉, 荣成博. 榆黄菇遗传多样性的ISSR和SRAP综合分析[J]. 生物技术通报, 2018, 34(4): 121-126. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||