








生物技术通报 ›› 2022, Vol. 38 ›› Issue (9): 198-206.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0510
孙威1( ), 张艳1, 王聿晗1, 徐僡1, 徐小蓉1(
), 张艳1, 王聿晗1, 徐僡1, 徐小蓉1( ), 鞠志刚2(
), 鞠志刚2( )
)
收稿日期:2022-04-25
出版日期:2022-09-26
发布日期:2022-10-11
作者简介:孙 威,女,博士,副教授,研究方向:植物生物技术;E-mail: 基金资助:
SUN Wei1( ), ZHANG Yan1, WANG Yu-han1, XU Hui1, XU Xiao-rong1(
), ZHANG Yan1, WANG Yu-han1, XU Hui1, XU Xiao-rong1( ), JU Zhi-gang2(
), JU Zhi-gang2( )
)
Received:2022-04-25
Published:2022-09-26
Online:2022-10-11
摘要:
类黄酮3-O-糖基转移酶(3GT)是花色素苷生物合成途径末端的酶,负责将糖基供体转移至花青素的3-OH位置,在增加花色素苷的稳定性与水溶性方面发挥重要作用。研究马缨杜鹃3GT在花色素苷生物合成中的功能,为马缨杜鹃花色形成及调控研究奠定基础。运用 RT-PCR 技术克隆获得马缨杜鹃3GT基因(Rd3GT1)全长CDS序列,并对其进行生物信息学分析,再利用DNA重组技术完成植物表达载体pBI121-Rd3GT1的构建,利用农杆菌介导法对矮牵牛进行遗传转化,同时对再生植株进行转基因及表型鉴定,并完成基因表达量及花色素苷检测分析。结果表明,Rd3GT1全长CDS序列为1 395 bp,编码464个氨基酸。蛋白多序列比对和系统进化分析表明,Rd3GT1属于3GT家族成员。与野生型相比,转Rd3GT1的矮牵牛植株花色由白色变为粉色,花色素苷与黄酮醇的积累显著增加,同时多个类黄酮合成相关基因表达显著升高。综上,Rd3GT1在花色素苷合成过程中发挥重要功能,可用于其它植物花色改良研究。
孙威, 张艳, 王聿晗, 徐僡, 徐小蓉, 鞠志刚. 马缨杜鹃Rd3GT1的克隆及对矮牵牛花色形成的影响[J]. 生物技术通报, 2022, 38(9): 198-206.
SUN Wei, ZHANG Yan, WANG Yu-han, XU Hui, XU Xiao-rong, JU Zhi-gang. Cloning of Rd3GT1 in Rhododendron delavayi and Its Effect on Flower Color Formation of Petunia hybrida[J]. Biotechnology Bulletin, 2022, 38(9): 198-206.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 引物用途Function of primer |
|---|---|---|
| Rd3GT-1 F1 | ACCAAACAAATACTGTAATAAT | 基因克隆 |
| Rd3GT-1 R1 | GATTACACCCATCTTTTATTCA | Gene cloning |
| Rd3GT-121F | GCTCTAGA ATGACCAAAAATATCTCA | 载体构建 |
| Rd3GT-121R | CGGGATCCCTAAAGATTGTACCCTGC | Vector construction |
| PhCHSA-F | GGCGCGATCATTATAGGTTC | 表达分析 |
| PhCHSA-R | TTTGAGATCAGCCCAGGAAC | Expression analysis |
| PhCHI-F | TACGGCGATAGGTGTGTATC | 表达分析 |
| PhCHI-R | GGCAAGATCGTAGTAACTCG | Expression analysis |
| PhF3H-F | ACTTGGATCACTGTTCAGCC | 表达分析 |
| PhF3H-R | ATACACTATCGCCTCTGGTG | Expression analysis |
| PhDFR-F | GCTATCATCTACGATGTGGC | 表达分析 |
| PhDFR-R | TGTCGACAAGTATCGATGGC | Expression analysis |
| PhANS-F | TACCTGAGACTGTCACTGAG | 表达分析 |
| PhANS-R | GCAGTATCCAGTTCATCCTC | Expression analysis |
| Ph3GT-F | GCAGTGGCAGAAGCATTAGA | 表达分析 |
| Ph3GT-R | CACATGATATGCCCTCCAAA | Expression analysis |
| PhAN11-F | GCCGCATTGCCGTGGGTAG | 表达分析 |
| PhAN11-R | GGGATTGGGTTTAGGGTTAGGGTTTC | Expression analysis |
| PhAN1-F | TCTGCCGGCGAATCAAATCAA | 表达分析 |
| PhAN1-R | GTCTGTACGCGGGCACTCTTAGC | Expression analysis |
| PhJAF13-F | ACGGATGATAATATGAGTAACGGTGTGC | 表达分析 |
| PhJAF13-R | CTTGATGGTCTAGTGGGGCAGGC | Expression analysis |
| PhAN2-F | GATGGACTTCAATGGTGGGCCAAT | 表达分析 |
| PhAN2-R | CGATGGTGCTGTTTCCTCATGCAA | Expression analysis |
| PhMYBx-F | GTGGCTCCTCGGATGTTAGTTTCA | 表达分析 |
| PhMYBx-R | GACCACCTCTCGCCAACCAAATTA | Expression analysis |
| PhActin2-F | CCTGATGAAGATCCTCACCGA | 表达分析 |
| PhActin2-R | CAAGAGCCACATAGGCAAGCT | Expression analysis |
表1 本文所用引物列表
Table 1 Primers used in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 引物用途Function of primer |
|---|---|---|
| Rd3GT-1 F1 | ACCAAACAAATACTGTAATAAT | 基因克隆 |
| Rd3GT-1 R1 | GATTACACCCATCTTTTATTCA | Gene cloning |
| Rd3GT-121F | GCTCTAGA ATGACCAAAAATATCTCA | 载体构建 |
| Rd3GT-121R | CGGGATCCCTAAAGATTGTACCCTGC | Vector construction |
| PhCHSA-F | GGCGCGATCATTATAGGTTC | 表达分析 |
| PhCHSA-R | TTTGAGATCAGCCCAGGAAC | Expression analysis |
| PhCHI-F | TACGGCGATAGGTGTGTATC | 表达分析 |
| PhCHI-R | GGCAAGATCGTAGTAACTCG | Expression analysis |
| PhF3H-F | ACTTGGATCACTGTTCAGCC | 表达分析 |
| PhF3H-R | ATACACTATCGCCTCTGGTG | Expression analysis |
| PhDFR-F | GCTATCATCTACGATGTGGC | 表达分析 |
| PhDFR-R | TGTCGACAAGTATCGATGGC | Expression analysis |
| PhANS-F | TACCTGAGACTGTCACTGAG | 表达分析 |
| PhANS-R | GCAGTATCCAGTTCATCCTC | Expression analysis |
| Ph3GT-F | GCAGTGGCAGAAGCATTAGA | 表达分析 |
| Ph3GT-R | CACATGATATGCCCTCCAAA | Expression analysis |
| PhAN11-F | GCCGCATTGCCGTGGGTAG | 表达分析 |
| PhAN11-R | GGGATTGGGTTTAGGGTTAGGGTTTC | Expression analysis |
| PhAN1-F | TCTGCCGGCGAATCAAATCAA | 表达分析 |
| PhAN1-R | GTCTGTACGCGGGCACTCTTAGC | Expression analysis |
| PhJAF13-F | ACGGATGATAATATGAGTAACGGTGTGC | 表达分析 |
| PhJAF13-R | CTTGATGGTCTAGTGGGGCAGGC | Expression analysis |
| PhAN2-F | GATGGACTTCAATGGTGGGCCAAT | 表达分析 |
| PhAN2-R | CGATGGTGCTGTTTCCTCATGCAA | Expression analysis |
| PhMYBx-F | GTGGCTCCTCGGATGTTAGTTTCA | 表达分析 |
| PhMYBx-R | GACCACCTCTCGCCAACCAAATTA | Expression analysis |
| PhActin2-F | CCTGATGAAGATCCTCACCGA | 表达分析 |
| PhActin2-R | CAAGAGCCACATAGGCAAGCT | Expression analysis |
| 培养基名称 Name of culture medium | 培养基配方 Ingredient culture |
|---|---|
| 共培养培养基 Cocultivation medium | MS基本培养基+BA(1 mg/L)+NAA(0.1 mg/L)+AS(20 mg/L) |
| 脱菌培养基 Bacteria-free medium | MS基本培养基+BA(1mg/L)+NAA(0.1 mg/L)+cef(400 mg/L) |
| 筛选培养基 Medium for screening | MS基本培养基+BA(1 mg/L)+NAA(0.1 mg/L)+cef(400 mg/L)+Kan(50 mg/L) |
| 生根培养基 Medium for growing roots | 1/2 MS基本培养基+NAA(0.3 mg/L)+cef(400 mg/L)+Kan(50 mg/L) |
表2 矮牵牛遗传转化培养基成分
Table 2 A variety of culture medium in the genetic transf-ormation experiment of P. hybrida
| 培养基名称 Name of culture medium | 培养基配方 Ingredient culture |
|---|---|
| 共培养培养基 Cocultivation medium | MS基本培养基+BA(1 mg/L)+NAA(0.1 mg/L)+AS(20 mg/L) |
| 脱菌培养基 Bacteria-free medium | MS基本培养基+BA(1mg/L)+NAA(0.1 mg/L)+cef(400 mg/L) |
| 筛选培养基 Medium for screening | MS基本培养基+BA(1 mg/L)+NAA(0.1 mg/L)+cef(400 mg/L)+Kan(50 mg/L) |
| 生根培养基 Medium for growing roots | 1/2 MS基本培养基+NAA(0.3 mg/L)+cef(400 mg/L)+Kan(50 mg/L) |
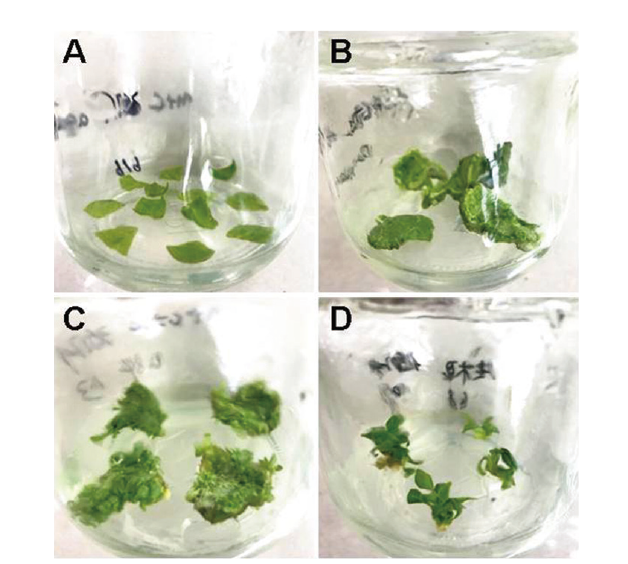
图6 矮牵牛的遗传转化过程 A:脱菌培养;B:筛选培养;C:继代培养;D:生根培养
Fig. 6 Genetic transformation process of P. hybrid A:Bacteria-free culture. B:Screening culture. C:Subculture. D:Rooting culture

图8 转基因花朵中花色素苷与黄酮醇成分分析 A, D:野生型;B, E:Rd3GT1-4转基因植株;C, F:Rd3GT1-5转基因植株
Fig. 8 Analysis of anthocyanin and flavonol components in transgenic flowers A, D:Wild type. B, E:Rd3GT1-4 transgenic. C, F:Rd3GT1-5 transgenic
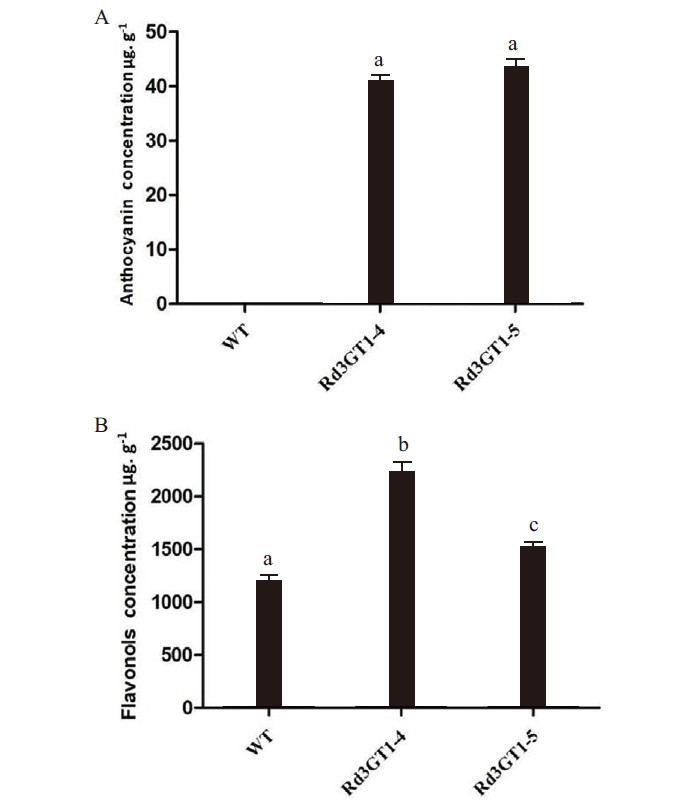
图9 转基因花朵中花色素苷与黄酮醇的定量分析 A:花色素苷;B:黄酮醇。a,b,c表示 0.01 水平上极显著差异
Fig. 9 Quantitative analysis of anthocyanin and flavonol in transgenic flowers A:Anthocyanin. B:Flavonol. a, b, c indicate very significant difference at the 0.01 level
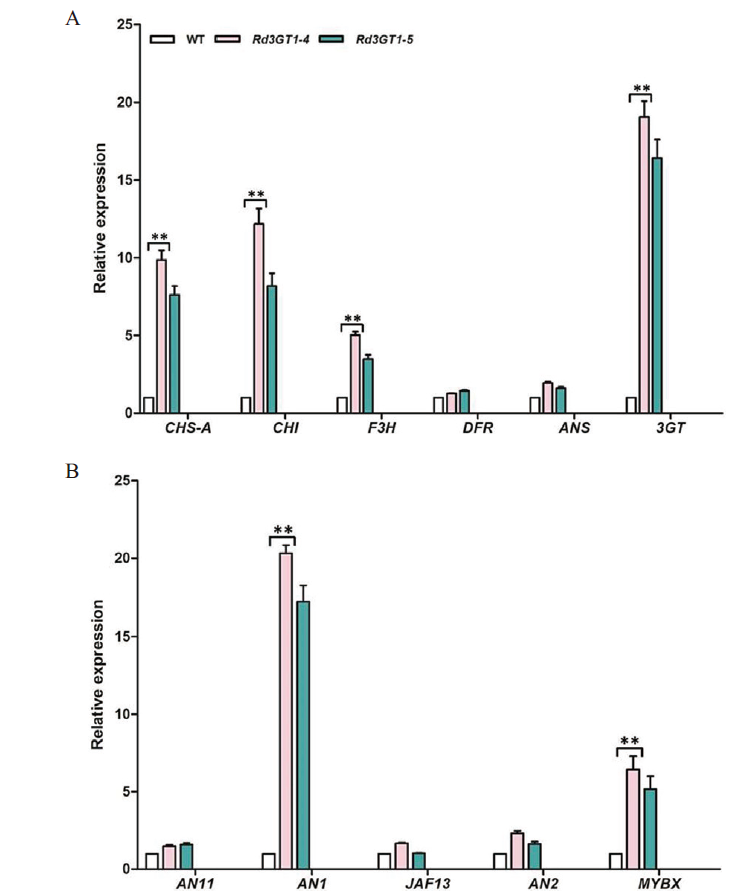
图10 转基因矮牵牛中类黄酮合成相关基因的表达分析 A:结构基因;B:转录因子;**表示 0.01 水平上极显著差异
Fig. 10 Expression profiles of flavonoid-related biosynth-etic genes in flowers of transgenic petunia A:Structure gene. B:Transcription factor. ** indicates very significant difference at the 0.01 level
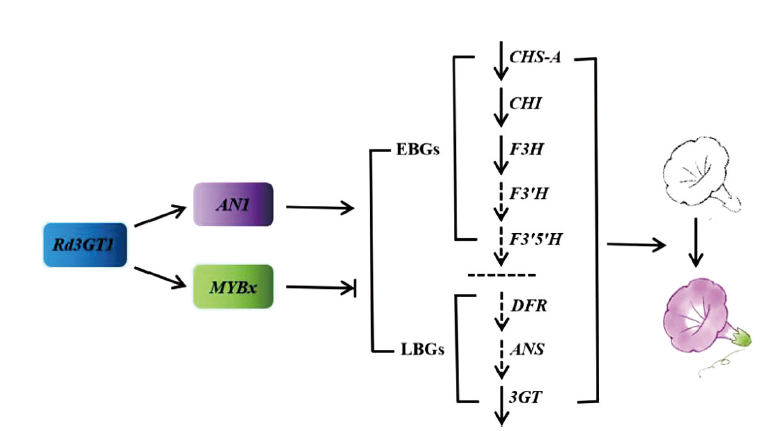
图11 Rd3GT1调节转基因矮牵牛花色的建议模式图 实线箭头表示基因表达量显著升高;虚线箭头表示基因表达量无显著变化
Fig. 11 Proposed model for regulation of transgenic petu-nia flower color by Rd3GT1 The solid arrows indicate the gene expression levels significantly increased. The dotted arrows indicate there is no significant change in gene expression levels
| [1] |
Falcone Ferreyra ML, Rius SP, Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications[J]. Front Plant Sci, 2012, 3: 222.
doi: 10.3389/fpls.2012.00222 pmid: 23060891 |
| [2] |
Forkmann G, Martens S. Metabolic engineering and applications of flavonoids[J]. Curr Opin Biotechnol, 2001, 12(2): 155-160.
doi: 10.1016/S0958-1669(00)00192-0 URL |
| [3] |
Li D, Chen G, Ma B, et al. Metabolic profiling and transcriptome analysis of mulberry leaves provide insights into flavonoid biosynthesis[J]. J Agric Food Chem, 2020, 68(5): 1494-1504.
doi: 10.1021/acs.jafc.9b06931 URL |
| [4] |
Sharma M, Cortes-Cruz M, Ahern KR, et al. Identification of the Pr1 gene product completes the anthocyanin biosynthesis pathway of maize[J]. Genetics, 2011, 188(1): 69-79.
doi: 10.1534/genetics.110.126136 pmid: 21385724 |
| [5] |
Katayama-Ikegami A, Byun Z, Okada S, et al. Characterization of the recombinant UDP: flavonoid 3-O-galactosyltransferase from Mangifera indica ‘Irwin'(MiUFGalT3)involved in skin coloring[J]. Hortic J, 2020, 89(5): 516-524.
doi: 10.2503/hortj.UTD-201 URL |
| [6] |
Tohge T, Nishiyama Y, Hirai MY, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor[J]. Plant J, 2005, 42(2): 218-235.
doi: 10.1111/j.1365-313X.2005.02371.x URL |
| [7] |
Yamazaki M, Yamagishi E, Gong ZZ, et al. Two flavonoid glucosyltransferases from Petunia hybrida: molecular cloning, biochemical properties and developmentally regulated expression[J]. Plant Mol Biol, 2002, 48(4): 401-411.
pmid: 11905966 |
| [8] | Sun W, Liang LJ, Meng XY, et al. Biochemical and molecular characterization of a flavonoid 3-O-glycosyltransferase responsible for anthocyanins and flavonols biosynthesis in Freesia hybrida[J]. Front Plant Sci, 2016, 7: 410. |
| [9] |
Wang XQ. Structure, mechanism and engineering of plant natural product glycosyltransferases[J]. FEBS Lett, 2009, 583(20): 3303-3309.
doi: 10.1016/j.febslet.2009.09.042 pmid: 19796637 |
| [10] |
le Roy J, Huss B, Creach A, et al. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants[J]. Front Plant Sci, 2016, 7: 735.
doi: 10.3389/fpls.2016.00735 pmid: 27303427 |
| [11] | 吴福建, 李凤兰, 黄凤兰, 等. 杜鹃花研究进展[J]. 东北农业大学学报, 2008, 39(1): 139-144. |
| Wu FJ, Li FL, Huang FL, et al. Research progress on Rhododendron[J]. J Northeast Agric Univ, 2008, 39(1): 139-144. | |
| [12] |
Griesbach RJ, Asen S, Leonnarat BA. Petunia hybrida anthocyanins acylated with caffeic acid[J]. Phytochemistry, 1991, 30(5): 1729-1731.
doi: 10.1016/0031-9422(91)84250-V URL |
| [13] |
Sun W, Meng XY, Liang LJ, et al. Overexpression of a Freesia hybrida flavonoid 3-O-glycosyltransferase gene, Fh3GT1, enhances transcription of key anthocyanin genes and accumulation of anthocyanin and flavonol in transgenic Petunia(Petunia hybrida)[J]. Vitro Cell Dev Biol Plant, 2017, 53(5): 478-488.
doi: 10.1007/s11627-017-9836-3 URL |
| [14] |
Downey MO, Harvey JS, Robinson SP. Analysis of tannins in seeds and skins of Shiraz grapes throughout berry development[J]. Aust J Grape Wine Res, 2003, 9(1): 15-27.
doi: 10.1111/j.1755-0238.2003.tb00228.x URL |
| [15] |
Griesbach RJ, Asen S. Characterization of the flavonol glycosides in Petunia[J]. Plant Sci, 1990, 70(1): 49-56.
doi: 10.1016/0168-9452(90)90031-I URL |
| [16] |
Fukuchi-Mizutani M, Okuhara H, Fukui Y, et al. Biochemical and molecular characterization of a novel UDP-glucose: anthocyanin 3'-O-glucosyltransferase, a key enzyme for blue anthocyanin biosynthesis, from Gentian[J]. Plant Physiol, 2003, 132(3): 1652-1663.
pmid: 12857844 |
| [17] |
Tanaka Y, Yonekura K, Fukuchi-Mizutani M, et al. Molecular and biochemical characterization of three anthocyanin synthetic enzymes from Gentiana triflora[J]. Plant Cell Physiol, 1996, 37(5): 711-716.
pmid: 8819318 |
| [18] |
Ford CM, Boss PK, Hoj PB. Cloning and characterization of Vitis vinifera UDP-glucose: flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo[J]. J Biol Chem, 1998, 273(15): 9224-9233.
doi: 10.1074/jbc.273.15.9224 pmid: 9535914 |
| [19] |
Modolo LV, Li LN, Pan HY, et al. Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of(Iso)flavonoids[J]. J Mol Biol, 2009, 392(5): 1292-1302.
doi: 10.1016/j.jmb.2009.08.017 pmid: 19683002 |
| [20] |
Davies K, Schwinn K, Deroles S, et al. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase[J]. Euphytica, 2003, 131: 259-268.
doi: 10.1023/A:1024018729349 URL |
| [21] |
Meyer P, Heidmann I, Forkmann G, et al. A new Petunia flower colour generated by transformation of a mutant with a maize gene[J]. Nature, 1987, 330(6149): 677-678.
doi: 10.1038/330677a0 URL |
| [22] |
Owens DK, Alerding AB, Crosby KC, et al. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis[J]. Plant Physiol, 2008, 147(3): 1046-1061.
doi: 10.1104/pp.108.117457 URL |
| [23] |
Gou JY, Felippes FF, Liu CJ, et al. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor[J]. Plant Cell, 2011, 23(4): 1512-1522.
doi: 10.1105/tpc.111.084525 URL |
| [24] | Wang CK, Chen PY, Wang HM, et al. Cosuppression of tobacco Chalcone synthase using Petunia Chalcone synthase constructs results in white flowers[J]. Bot Stud, 2006, 47: 71-82. |
| [25] |
Nishihara M, Nakatsuka T, Yamamura S. Flavonoid components and flower color change in transgenic tobacco plants by suppression of Chalcone isomerase gene[J]. FEBS Lett, 2005, 579(27): 6074-6078.
pmid: 16226261 |
| [26] |
Forkmann G, Stotz G. Selection and characterisation of flavanone 3-hydroxylase mutants of Dahlia, Streptocarpus, Verbena and Zinnia[J]. Planta, 1984, 161(3): 261-265.
doi: 10.1007/BF00982923 pmid: 24253654 |
| [27] |
Zuker A, Tzfira T, Ben-Meir H, et al. Modification of flower color and fragrance by antisense suppression of the flavanone 3-hydroxylase gene[J]. Molecular Breeding, 2002, 9:33-41.
doi: 10.1023/A:1019204531262 URL |
| [28] |
Zhao ZC, Hu GB, Hu FC, et al. The UDP glucose: flavonoid-3-O-glucosyltransferase(UFGT)gene regulates anthocyanin biosynthesis in Litchi(Litchi chinesis Sonn.)during fruit coloration[J]. Mol Biol Rep, 2012, 39(6): 6409-6415.
doi: 10.1007/s11033-011-1303-3 URL |
| [29] |
Aharoni A, de Vos CH, Wein M, et al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco[J]. Plant J, 2001, 28(3): 319-332.
pmid: 11722774 |
| [30] |
Nakatsuka T, Yamada E, Saito M, et al. Heterologous expression of Gentian MYB1R transcription factors suppresses anthocyanin pigmentation in tobacco flowers[J]. Plant Cell Rep, 2013, 32(12): 1925-1937.
doi: 10.1007/s00299-013-1504-4 pmid: 24037114 |
| [1] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [2] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [3] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [4] | 郭三保, 宋美玲, 李灵心, 尧子钊, 桂明明, 黄胜和. 斑地锦查尔酮合酶基因及启动子的克隆与分析[J]. 生物技术通报, 2023, 39(4): 148-156. |
| [5] | 杨俊钊, 张新蕊, 赵国柱, 郑菲. 新型GH5家族多结构域纤维素酶的结构与功能研究[J]. 生物技术通报, 2023, 39(4): 71-80. |
| [6] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [7] | 陈楚雯, 李洁, 赵瑞鹏, 刘媛, 吴锦波, 李志雄. 藏鸡GPX3基因的克隆、组织表达谱研究及功能预测[J]. 生物技术通报, 2023, 39(3): 311-320. |
| [8] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [9] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [10] | 侯瑞泽, 鲍悦, 陈启亮, 毛桂玲, 韦博霖, 侯雷平, 李梅兰. 普通白菜PRR5的克隆、表达及功能验证[J]. 生物技术通报, 2023, 39(10): 128-135. |
| [11] | 杨敏, 龙雨青, 曾娟, 曾梅, 周新茹, 王玲, 付学森, 周日宝, 刘湘丹. 灰毡毛忍冬UGTPg17、UGTPg36基因克隆及功能研究[J]. 生物技术通报, 2023, 39(10): 256-267. |
| [12] | 郭文博, 路杨, 隋丽, 赵宇, 邹晓威, 张正坤, 李启云. 球孢白僵菌真菌病毒BbPmV-4外壳蛋白多克隆抗体制备及应用[J]. 生物技术通报, 2023, 39(10): 58-67. |
| [13] | 周家燕, 邹建, 陈卫英, 吴一超, 陈奚潼, 王倩, 曾文静, 胡楠, 杨军. 植物多基因干扰载体体系构建与效用分析[J]. 生物技术通报, 2023, 39(1): 115-126. |
| [14] | 陈光, 李佳, 杜瑞英, 王旭. 水稻盐敏感突变体ss2的鉴定与基因功能分析[J]. 生物技术通报, 2022, 38(9): 158-166. |
| [15] | 李秀青, 胡子曜, 雷建峰, 代培红, 刘超, 邓嘉辉, 刘敏, 孙玲, 刘晓东, 李月. 棉花黄萎病抗性相关基因GhTIFY9的克隆与功能分析[J]. 生物技术通报, 2022, 38(8): 127-134. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||