








生物技术通报 ›› 2023, Vol. 39 ›› Issue (9): 268-280.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0280
薛宁1,2,3( ), 王瑾2,3, 李世新1,2,3, 刘叶2,3, 程海娇2,3, 张玥2,3, 毛雨丰2,3, 王猛2,3(
), 王瑾2,3, 李世新1,2,3, 刘叶2,3, 程海娇2,3, 张玥2,3, 毛雨丰2,3, 王猛2,3( )
)
收稿日期:2023-03-27
出版日期:2023-09-26
发布日期:2023-10-24
通讯作者:
王猛,男,研究员,博士生导师,研究方向:合成生物学和高通量自动化;E-mail: wangmeng@tib.cas.cn作者简介:薛宁,女,硕士研究生,研究方向:生物工程;E-mail: xuening@tib.cas.cn王瑾同为本文第一作者
基金资助:
XUE Ning1,2,3( ), WANG Jin2,3, LI Shi-xin1,2,3, LIU Ye2,3, CHENG Hai-jiao2,3, ZHANG Yue2,3, MAO Yu-feng2,3, WANG Meng2,3(
), WANG Jin2,3, LI Shi-xin1,2,3, LIU Ye2,3, CHENG Hai-jiao2,3, ZHANG Yue2,3, MAO Yu-feng2,3, WANG Meng2,3( )
)
Received:2023-03-27
Published:2023-09-26
Online:2023-10-24
摘要:
L-苯丙氨酸(L-Phe)是重要的食品及医药中间体,但由于其生物合成途径较长且调控机制复杂,仅依靠质粒或者周期漫长的基因组逐轮改造,难以实现各个模块之间的通量平衡,在一定程度上限制了其产量的进一步提升。本研究将L-Phe生物合成途径中的7个携带组成型启动子PF11及核糖体结合位点(ribosome binding site, RBS)为GGGGGGGG的关键基因ppsA、tktA、aroFfbr、aroE、aroL、pheAfbr和tyrB,整合到谷氨酸棒杆菌(Corynebacterium glutamicum)的基因组中;利用课题组前期研发的基于CRISPR/Cas系统和胞苷脱氨酶的无模板基因表达调控技术(base editor-targeted and template-free expression regulation, BETTER),对每个关键基因的RBS进行同步编辑,生成富含G/A的RBS突变文库;利用前期开发的荧光蛋白型L-Phe生物传感器结合液滴微流控系统(droplet microfluidics)对RBS突变文库进行高通量筛选。最终,从大约7万个RBS突变文库中筛选到4株L-Phe产量提升不同程度的突变菌株,其72 h摇瓶发酵产量分别为2.06、1.30、4.42和7.44 mmol/L,相比对照菌株C.g-2(0.6 mmol/L)分别提高了2.43、1.17、6.37、11.40倍。利用碱基编辑器同时调节多基因的表达水平并筛选组合文库,可以为L-Phe工程育种提供一种可行策略。
薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280.
XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening[J]. Biotechnology Bulletin, 2023, 39(9): 268-280.
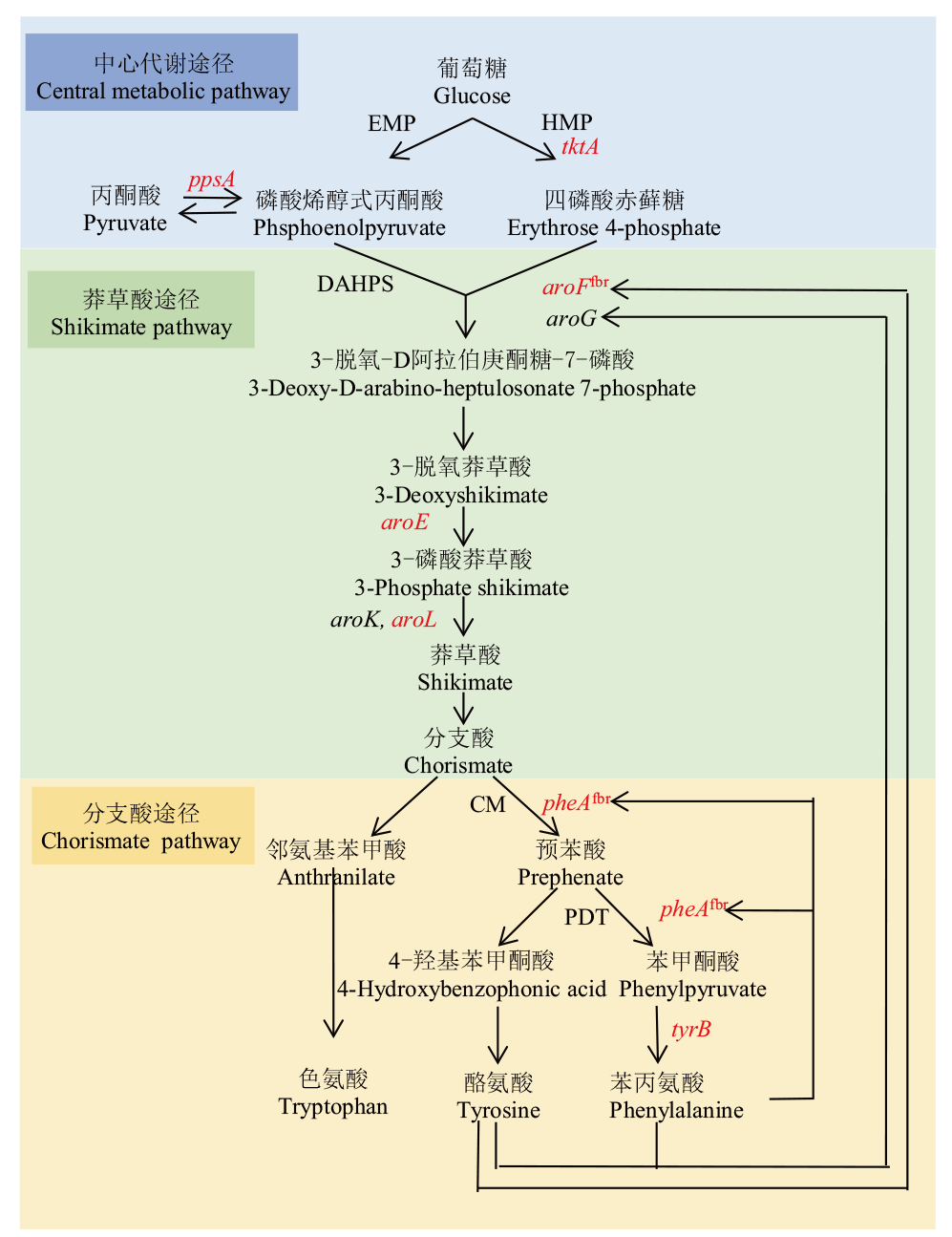
图1 谷氨酸棒杆菌中L-Phe生物合成途径 红色:本研究选择基因;EMP:糖酵解途径;HMP:磷酸戊糖途径;DAHPS:3-脱氧-D-阿拉伯庚酮糖酸7-磷酸合成酶;CM:分支酸变位酶;PDT:预苯酸脱水酶
Fig. 1 L-phenylalanine biosynthesis pathway in C. glutam-icum Red: Selected genes for this study. EMP: Embden-Meyerhof-Parnas pathway. HMP: Phophoenol pyruvate. DAHPS: 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase. CM: Feedback-inhibition resistance chorsmate mutase. PDT: Prephenate dehydratase
| Strain/ Plasmid | 名称 Name | 特征 Characteristics | 来源/文献 Source/Reference |
|---|---|---|---|
| Strain | E. coli DH5α | Host for plasmid cloning | Lab stock |
| E. coli MG1655 | Wild-type strain | Lab stock | |
| C. glutamicum ATCC 13032 | Wild-type strain | Lab stock | |
| C.g-0 | C. glutamicum ATCC 13032, ΔCGP2::pheAfbr-aroFfbr | Lab stock | |
| C.g-1 | C.g-0, ΔaroP::tktA-ppsA | This study | |
| C.g-2 | C.g-1, ΔldhA::aroE-aroL-tyrB | This study | |
| C.g-3 | C.g-2, pgRNA-RBS2 and pXMJ19-nCas9-AID | This study | |
| C.g-2-aroE-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-aroE-gfp | This study | |
| C.g-2-aroL-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-aroL-gfp | This study | |
| C.g-2-tyrB-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-tyrB-gfp | This study | |
| C.g-2-tktA-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-tktA-gfp | This study | |
| C.g-2-ppsA-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-ppsA-gfp | This study | |
| C.g-2-pheAfbr-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-pheAfbr-gfp | This study | |
| C.g-2-aroFfbr-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-aroFfbr-gfp | This study | |
| C.g-Mut4-aroE-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-aroE-gfp | This study | |
| C.g-Mut4-aroL-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g- Mut4-aroL-gfp | This study | |
| C.g-Mut4-tyrB-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-tyrB-gfp | This study | |
| C.g-Mut4-tktA-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-tktA-gfp | This study | |
| C.g-Mut4-ppsA-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-ppsA-gfp | This study | |
| C.g-Mut4-pheAfbr-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-pheAfbr-gfp | This study | |
| C.g-Mut4-aroFfbr-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-aroFfbr-gfp | This study | |
| Plasmid | pgRNA-RBS2 | pgRNA-ccdBNoPer derivative expressing two gRNAs(N20: 5'-CCCCCCCCTAGCTCTAAAAC-3'; N20: 5'-CCCCCTAGCTCTAAAACAGG-3')targeting the GGGGGGGG RBS | [ |
| pXMJ19-nCas9-AID | pXMJ19 carrying nCas9(D10A)-AID cassette driven by IPTG-inducible promoter Ptac, CmR, temperature-sensitive | [ | |
| pK18mobsacB | Gene deletion/integration vector, mob, sacB, KmR | Lab stock | |
| pk18-ΔCGP2::dxs-sxr-ispD | Plasmids derivative for integrating artificial dxs-sxr-ispD cluster preloaded with GGGGGGGG RBSs, promoter P11F and terminator rrnB at CGP2 locus resistance | Lab stock | |
| pk18-ΔldhA::gfp | Plasmids derivative for integrating artificial gfp cluster preloaded with GGGGGGGG RBSs, promoter P11F and terminator rrnB at ldhA locus | Lab stock | |
| pK18-ΔaroP::tktA-ppsA | Plasmids derivative for integrating artificial tktA-ppsA cluster preloaded with GGGGGGGG RBSs, promoter P11F and terminator rrnB at aroP locus | This study | |
| pK18-ΔldhA::aroE-aroL-tyrB | Plasmids derivative for integrating artificial aroE-aroL-tyrB cluster preloaded with GGGGGGGG RBSs, promoter P11F and terminator rrnB at ldhA locus | This study | |
| pEC-XK99E-C.g-2-aroE-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-aroL-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-tyrB-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-tktA-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-ppsA-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-pheAfbr-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-aroFfbr-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-aroE-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-aroL-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-tyrB-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-tktA-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-ppsA-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-pheAfbr-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-aroFfbr-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study |
表1 本研究用到的菌株和质粒
Table 1 Strains and plasmids used in this study
| Strain/ Plasmid | 名称 Name | 特征 Characteristics | 来源/文献 Source/Reference |
|---|---|---|---|
| Strain | E. coli DH5α | Host for plasmid cloning | Lab stock |
| E. coli MG1655 | Wild-type strain | Lab stock | |
| C. glutamicum ATCC 13032 | Wild-type strain | Lab stock | |
| C.g-0 | C. glutamicum ATCC 13032, ΔCGP2::pheAfbr-aroFfbr | Lab stock | |
| C.g-1 | C.g-0, ΔaroP::tktA-ppsA | This study | |
| C.g-2 | C.g-1, ΔldhA::aroE-aroL-tyrB | This study | |
| C.g-3 | C.g-2, pgRNA-RBS2 and pXMJ19-nCas9-AID | This study | |
| C.g-2-aroE-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-aroE-gfp | This study | |
| C.g-2-aroL-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-aroL-gfp | This study | |
| C.g-2-tyrB-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-tyrB-gfp | This study | |
| C.g-2-tktA-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-tktA-gfp | This study | |
| C.g-2-ppsA-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-ppsA-gfp | This study | |
| C.g-2-pheAfbr-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-pheAfbr-gfp | This study | |
| C.g-2-aroFfbr-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-2-aroFfbr-gfp | This study | |
| C.g-Mut4-aroE-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-aroE-gfp | This study | |
| C.g-Mut4-aroL-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g- Mut4-aroL-gfp | This study | |
| C.g-Mut4-tyrB-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-tyrB-gfp | This study | |
| C.g-Mut4-tktA-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-tktA-gfp | This study | |
| C.g-Mut4-ppsA-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-ppsA-gfp | This study | |
| C.g-Mut4-pheAfbr-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-pheAfbr-gfp | This study | |
| C.g-Mut4-aroFfbr-gfp | C. glutamicum haboring plasmid pEC-XK99E-C.g-Mut4-aroFfbr-gfp | This study | |
| Plasmid | pgRNA-RBS2 | pgRNA-ccdBNoPer derivative expressing two gRNAs(N20: 5'-CCCCCCCCTAGCTCTAAAAC-3'; N20: 5'-CCCCCTAGCTCTAAAACAGG-3')targeting the GGGGGGGG RBS | [ |
| pXMJ19-nCas9-AID | pXMJ19 carrying nCas9(D10A)-AID cassette driven by IPTG-inducible promoter Ptac, CmR, temperature-sensitive | [ | |
| pK18mobsacB | Gene deletion/integration vector, mob, sacB, KmR | Lab stock | |
| pk18-ΔCGP2::dxs-sxr-ispD | Plasmids derivative for integrating artificial dxs-sxr-ispD cluster preloaded with GGGGGGGG RBSs, promoter P11F and terminator rrnB at CGP2 locus resistance | Lab stock | |
| pk18-ΔldhA::gfp | Plasmids derivative for integrating artificial gfp cluster preloaded with GGGGGGGG RBSs, promoter P11F and terminator rrnB at ldhA locus | Lab stock | |
| pK18-ΔaroP::tktA-ppsA | Plasmids derivative for integrating artificial tktA-ppsA cluster preloaded with GGGGGGGG RBSs, promoter P11F and terminator rrnB at aroP locus | This study | |
| pK18-ΔldhA::aroE-aroL-tyrB | Plasmids derivative for integrating artificial aroE-aroL-tyrB cluster preloaded with GGGGGGGG RBSs, promoter P11F and terminator rrnB at ldhA locus | This study | |
| pEC-XK99E-C.g-2-aroE-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-aroL-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-tyrB-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-tktA-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-ppsA-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-pheAfbr-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-2-aroFfbr-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-aroE-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-aroL-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-tyrB-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-tktA-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-ppsA-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-pheAfbr-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study | |
| pEC-XK99E-C.g-Mut4-aroFfbr-gfp | Plasmids carrying RBS strength reporter(gfp)cassette driven by promoter P11F, KmR | This study |
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| 1-AroP-F | ATGGACGGGACCATCCTGGA |
| 1-AroP-R | TGAATTACACTGTACCTGTTGCGTCGCAACATTGCCAGCTTATGTGGAGAAAACAGTCCGGCACCAATTGC |
| TktA-F | ACAGGTACAGTGTAATTCAGGAGAAAATCGCACCGGACCTCCTGTTTTAGAGCTAGGGGGGGGTTGAGAATGGACACCAAGGCTGTAGA |
| TktA-R | TTAACCGTTAATGGAGTCCTTGGC |
| 1-rrnB-P11F-F | AGGACTCCATTAACGGTTAAGGCTGTTTTGGCGGATGAGA |
| 1-rrnB-P11F-R | GCTGGACAAACGGGATGTTCATTCTCAACCCCCCCCTAGCTC |
| PpsA-F | ATGAACATCCCGTTTGTCCAGC |
| PpsA-R | TTACTTCGTGCCGGTCATTGC |
| rrnB-PpsA-F | CAATGACCGGCACGAAGTAAGGCTGTTTTGGCGGATGAGA |
| rrnB-PpsA-R | AAGAATCCGTACACAATTGCAGAGTTTGTAGAAACGCAAAAAGGC |
| 2-AroP-F | GCAATTGTGTACGGATTCTTGGT |
| 2-AroP-R | GTTGGAGTGCAAAGCTGTGTTG |
| pk18-ΔAroP-F | ACACAGCTTTGCACTCCAACGGATCCTCTAGAGTCGACCTGC |
| pk18-ΔAroP-R | TCCAGGATGGTCCCGTCCATGGATCCCCGGGTACCGAG |
| AroE-F | CAGGAGAAAATCGCACCGGACCTCCTGTTTTAGAGCTAGGGGGGGGTTGAGAATGAACGACAGTATTCTCCTCGGC |
| AroE-R | TTAGAGGGACAGGAAAGTTTCCCGC |
| AroL-F | AAACTTTCCTGTCCCTCTAACCTCCTGTTTTAGAGCTAGGGGGGGGTTGAGAATGACACAACCTCTTTTTCTGATCG |
| AroL-R | TCAACAATTGATCGTCTGTGCCAGG |
| 2-rrnB-P11F-F | CACAGACGATCAATTGTTGAGGCTGTTTTGGCGGATGAGA |
| 2-rrnB-P11F-R | GCGTCAACTTTTTGAAACATTCTCAACCCCCCCCTAGCTCTAAA |
| TyrB-F | ATGTTTCAAAAAGTTGACGCCTACG |
| TyrB-R | TTACATCACCGCAGCAAACG |
| rrnB-TyrB-F | CGTTTGCTGCGGTGATGTAAGGCTGTTTTGGCGGATGAGA |
| rrnB-TyrB-R | TTTTGTAGGATTGCGCGGGTAGAGTTTGTAGAAACGCAAAAAGGC |
| pk18-ΔLdhA-F1 | ACCCGCGCAATCCTACAAAAC |
| pk18-ΔLdhA-R1 | TTCTTCGCCTTGGTAGCCATCTTC |
| pk18-ΔLdhA-F2 | GAAGATGGCTACCAAGGCGAAGAA |
| PheA-AroF-JYZ-F1 | TTCCAGAACCAGATAATCCGCAT |
| PheA-AroF-JYZ-R1 | ATCTGGGCTTCTTGTTGCAGG |
| PheA-AroF-JYZ-F2 | GTGTCTGATCAGGTTCCGGC |
| PheA-AroF-YJZ-R2 | TCGCTGCCATACACTTTTTCAAG |
| pk18-ΔLdhA-R2 | TCCGGTGCGATTTTCTCCTG |
| F1 | GGTAGATTAAGGTTCCGAAGGGCA |
| R1 | TTAACCGTTAATGGAGTCCTTGGC |
| F2 | GCTGTGACCGCTCGTGTG |
| R2-1 | GGATTGGGCCGATGGATGTG |
| R2-2 | ATCGCCATTCGCCATTCAGG |
| F3 | AATTGTCACGCAGTTTCCGCC |
| R3 | TAAATAAAGCGAAGCGCCATGAGG |
| F4 | ATGTTTCAAAAAGTTGACGCCTACG |
| R4-1 | ACTTTGAGGTTGATGCGCTGAAC |
| R4-2 | CGAAAGGGGGATGTGCTGCAA |
| AroE-AroL-TyrB-RBS-F | TTGGTTTTGCTCGATTTACAATGTGAT |
| AroE-AroL-TyrB-RBS-R | AATCCGCGCCCACTTTCAAT |
| TktA-PpsA-RBS-F | GCAACTTTGGTGTCAGCTCCG |
| TktA-PpsA-RBS-R | CCACCGCATCGTTCCATGAG |
| PheA-AroF-RBS-F | CTGCGGCAAGGGCTTTAAAT |
| PheA-AroF-RBS-R | ATAAAGACGGCAGGACTTCTTGG |
表2 本研究用到的引物
Table 2 Primers and plasmids used in this study
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| 1-AroP-F | ATGGACGGGACCATCCTGGA |
| 1-AroP-R | TGAATTACACTGTACCTGTTGCGTCGCAACATTGCCAGCTTATGTGGAGAAAACAGTCCGGCACCAATTGC |
| TktA-F | ACAGGTACAGTGTAATTCAGGAGAAAATCGCACCGGACCTCCTGTTTTAGAGCTAGGGGGGGGTTGAGAATGGACACCAAGGCTGTAGA |
| TktA-R | TTAACCGTTAATGGAGTCCTTGGC |
| 1-rrnB-P11F-F | AGGACTCCATTAACGGTTAAGGCTGTTTTGGCGGATGAGA |
| 1-rrnB-P11F-R | GCTGGACAAACGGGATGTTCATTCTCAACCCCCCCCTAGCTC |
| PpsA-F | ATGAACATCCCGTTTGTCCAGC |
| PpsA-R | TTACTTCGTGCCGGTCATTGC |
| rrnB-PpsA-F | CAATGACCGGCACGAAGTAAGGCTGTTTTGGCGGATGAGA |
| rrnB-PpsA-R | AAGAATCCGTACACAATTGCAGAGTTTGTAGAAACGCAAAAAGGC |
| 2-AroP-F | GCAATTGTGTACGGATTCTTGGT |
| 2-AroP-R | GTTGGAGTGCAAAGCTGTGTTG |
| pk18-ΔAroP-F | ACACAGCTTTGCACTCCAACGGATCCTCTAGAGTCGACCTGC |
| pk18-ΔAroP-R | TCCAGGATGGTCCCGTCCATGGATCCCCGGGTACCGAG |
| AroE-F | CAGGAGAAAATCGCACCGGACCTCCTGTTTTAGAGCTAGGGGGGGGTTGAGAATGAACGACAGTATTCTCCTCGGC |
| AroE-R | TTAGAGGGACAGGAAAGTTTCCCGC |
| AroL-F | AAACTTTCCTGTCCCTCTAACCTCCTGTTTTAGAGCTAGGGGGGGGTTGAGAATGACACAACCTCTTTTTCTGATCG |
| AroL-R | TCAACAATTGATCGTCTGTGCCAGG |
| 2-rrnB-P11F-F | CACAGACGATCAATTGTTGAGGCTGTTTTGGCGGATGAGA |
| 2-rrnB-P11F-R | GCGTCAACTTTTTGAAACATTCTCAACCCCCCCCTAGCTCTAAA |
| TyrB-F | ATGTTTCAAAAAGTTGACGCCTACG |
| TyrB-R | TTACATCACCGCAGCAAACG |
| rrnB-TyrB-F | CGTTTGCTGCGGTGATGTAAGGCTGTTTTGGCGGATGAGA |
| rrnB-TyrB-R | TTTTGTAGGATTGCGCGGGTAGAGTTTGTAGAAACGCAAAAAGGC |
| pk18-ΔLdhA-F1 | ACCCGCGCAATCCTACAAAAC |
| pk18-ΔLdhA-R1 | TTCTTCGCCTTGGTAGCCATCTTC |
| pk18-ΔLdhA-F2 | GAAGATGGCTACCAAGGCGAAGAA |
| PheA-AroF-JYZ-F1 | TTCCAGAACCAGATAATCCGCAT |
| PheA-AroF-JYZ-R1 | ATCTGGGCTTCTTGTTGCAGG |
| PheA-AroF-JYZ-F2 | GTGTCTGATCAGGTTCCGGC |
| PheA-AroF-YJZ-R2 | TCGCTGCCATACACTTTTTCAAG |
| pk18-ΔLdhA-R2 | TCCGGTGCGATTTTCTCCTG |
| F1 | GGTAGATTAAGGTTCCGAAGGGCA |
| R1 | TTAACCGTTAATGGAGTCCTTGGC |
| F2 | GCTGTGACCGCTCGTGTG |
| R2-1 | GGATTGGGCCGATGGATGTG |
| R2-2 | ATCGCCATTCGCCATTCAGG |
| F3 | AATTGTCACGCAGTTTCCGCC |
| R3 | TAAATAAAGCGAAGCGCCATGAGG |
| F4 | ATGTTTCAAAAAGTTGACGCCTACG |
| R4-1 | ACTTTGAGGTTGATGCGCTGAAC |
| R4-2 | CGAAAGGGGGATGTGCTGCAA |
| AroE-AroL-TyrB-RBS-F | TTGGTTTTGCTCGATTTACAATGTGAT |
| AroE-AroL-TyrB-RBS-R | AATCCGCGCCCACTTTCAAT |
| TktA-PpsA-RBS-F | GCAACTTTGGTGTCAGCTCCG |
| TktA-PpsA-RBS-R | CCACCGCATCGTTCCATGAG |
| PheA-AroF-RBS-F | CTGCGGCAAGGGCTTTAAAT |
| PheA-AroF-RBS-R | ATAAAGACGGCAGGACTTCTTGG |
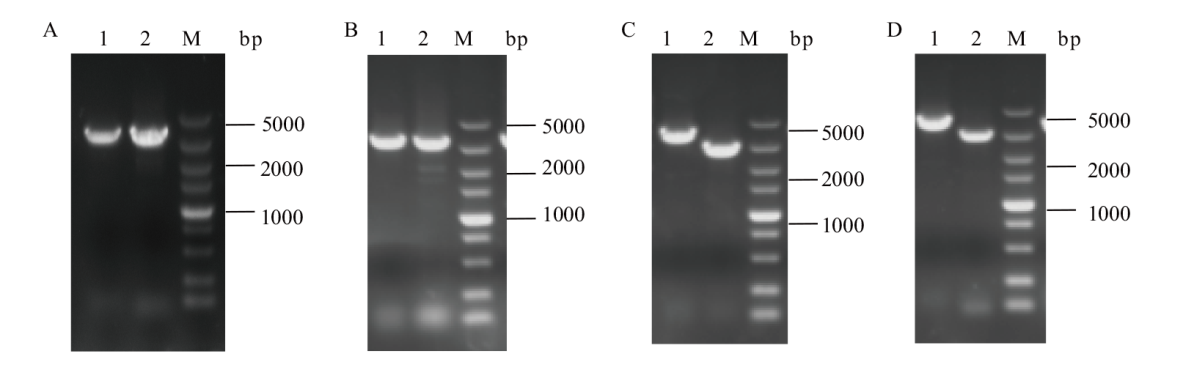
图3 C.g-1和C.g-2重组菌株两轮单交换验证 A:C.g-1第一轮单交换验证,使用引物F1/R1、F2/R2-2验证;B:C.g-1第二轮单交换验证,使用引物F1/R1、F2/R2-1验证;C:C.g-2第一轮单交换验证,使用引物F3/R3、F4/R4-2验证;D:C.g-2第二轮单交换验证,使用引物F3/R3、F4/R4-1验证。1和2分别为整合基因的上下两个片段,M为5 000 bp DNA marker
Fig. 3 PCR identification of the first-cross and second-cross of recombinant strain C.g-1 and C.g-2 A: PCR identification of the first-cross of recombinant strain C.g-1, validation using primer F1/R1, F2/R2-2. B: PCR identification of the second-cross of recombinant strain C.g-1, validation using primer F1/R1, F2/R2-1. C: PCR identification of the first-cross of recombinant strain C.g-2, validation using primers F3/R3, F4/R4-2. D: PCR identification of the second-cross of recombinant strain C.g-2, validation using primers F3/R3, F4/R4-1. 1 and 2 are the upper and lower segments of the integration gene, M is 5 000 bp DNA marker

图5 液滴收集、培养与观察及液滴分选 A:液滴收集和培养;B:液滴培养与观察(B:白场,F荧光场,比例尺:20 µm);C:液滴分选;D:液滴分选与分选打点
Fig. 5 Droplet collection, culture and observation, and droplet sorting A: Droplet collection and culture. B: Droplet culture and observation(B: white field, F: fluorescence field, scale bar: 20 μm). C: Droplet sorting. D: Droplet sorting and sorting points
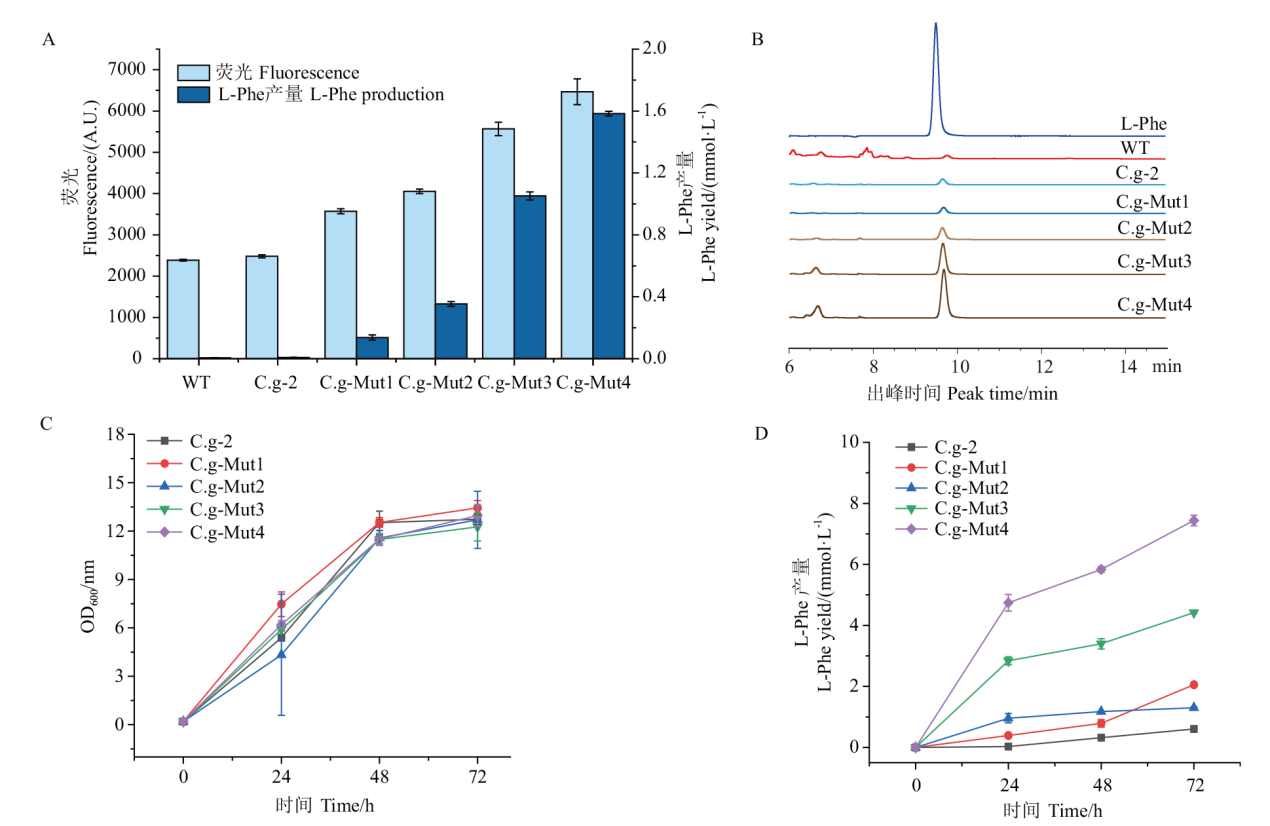
图6 生物传感器及高效液相色谱检测L-Phe产量 A:菌株孔板发酵荧光测定及高效液相色谱测定L-Phe产量;B:样品出锋时间;C:菌株摇瓶生长曲线;D:菌株产L-Phe摇瓶发酵曲线
Fig. 6 Biosensor and high performance liquid chromatography for L-Phe yield A: Fluorescence determination of strain plate fermentation and determination of L-Phe yield by high performance liquid chromatography. B: Sample strike time. C: Growth curve of strain in shaking flask. D: Fermentation curve of L-Phe in shaking flask produced by strain
| 基因 Gene | C.g-Mut1 | C.g-Mut2 | C.g-Mut3 | C.g-Mut4 |
|---|---|---|---|---|
| aroE | AAAAGGGG | GGAAAGGG | GGGGGGGG | GGAGGGGG |
| aroL | GGGGGGGG | AAAAAGGG | AAAAAGGG | GAAGGGGG |
| tyrB | GGGGGGGG | GAAAGGGG | GGAGGGGG | AAAAAGGG |
| tktA | GGGGGGAG | GAAAAAAG | AAAAAGGG | GGAGGGGG |
| ppsA | GGGAGGGG | ------ | ------ | GAAAGGGG |
| pheAfbr | GAAAGGGG | GGAAAGAG | GAAAGAAA | GGAAAAAA |
| aroFfbr | GAAAGGGG | AAAAAGAG | GAAAAGAG | GAAAGGAA |
表3 突变菌株RBS测序
Table 3 RBS sequencing for mutated strains
| 基因 Gene | C.g-Mut1 | C.g-Mut2 | C.g-Mut3 | C.g-Mut4 |
|---|---|---|---|---|
| aroE | AAAAGGGG | GGAAAGGG | GGGGGGGG | GGAGGGGG |
| aroL | GGGGGGGG | AAAAAGGG | AAAAAGGG | GAAGGGGG |
| tyrB | GGGGGGGG | GAAAGGGG | GGAGGGGG | AAAAAGGG |
| tktA | GGGGGGAG | GAAAAAAG | AAAAAGGG | GGAGGGGG |
| ppsA | GGGAGGGG | ------ | ------ | GAAAGGGG |
| pheAfbr | GAAAGGGG | GGAAAGAG | GAAAGAAA | GGAAAAAA |
| aroFfbr | GAAAGGGG | AAAAAGAG | GAAAAGAG | GAAAGGAA |
| [1] |
Liu YF, Xu YR, Ding DQ, et al. Genetic engineering of Escherichia coli to improve L-phenylalanine production[J]. BMC Biotechnol, 2018, 18(1): 5.
doi: 10.1186/s12896-018-0418-1 |
| [2] |
Yuan PP, Cao WJ, Wang Z, et al. Enhancement of L-phenylalanine production by engineered Escherichia coli using phased exponential L-tyrosine feeding combined with nitrogen source optimization[J]. J Biosci Bioeng, 2015, 120(1): 36-40.
doi: 10.1016/j.jbiosc.2014.12.002 URL |
| [3] | 齐欣, 赵温涛, 聂建明, 等. DL-苯丙氨酸合成新方法[J]. 化学工业与工程, 2002, 19(5): 394-397. |
|
Qi X, Zhao WT, Nie JM, et al. A new method of preparing DL-phenylalanine[J]. Chem Ind Eng, 2002, 19(5):394-397.
doi: 10.1021/ie50207a020 URL |
|
| [4] | 李永辉, 刘云, 徐琪寿. 苯丙氨酸生物合成的研究进展[J]. 生物技术通讯, 2002, 13(4): 296-300. |
| Li YH, Liu Y, Xu QS. Advances in biosynthesis of phenylalanine[J]. Lett Biotechnol, 2002, 13(4): 296-300. | |
| [5] |
Báez-Viveros JL, Osuna J, Hernández-Chávez G, et al. Metabolic engineering and protein directed evolution increase the yield of L-phenylalanine synthesized from glucose in Escherichia coli[J]. Biotechnol Bioeng, 2004, 87(4): 516-524.
doi: 10.1002/bit.20159 pmid: 15286989 |
| [6] |
Chen ML, Liang HY, Han C, et al. Engineering of global transcription factor FruR to redirect the carbon flow in Escherichia coli for enhancing L-phenylalanine biosynthesis[J]. Microb Cell Fact, 2022, 21(1): 222.
doi: 10.1186/s12934-022-01954-7 |
| [7] |
Zhou HY, Liao XY, Wang TW, et al. Enhanced l-phenylalanine biosynthesis by co-expression of pheAfbr and aroFwt[J]. Bioresour Technol, 2010, 101(11): 4151-4156.
doi: 10.1016/j.biortech.2010.01.043 URL |
| [8] | 门佳轩, 熊博, 郝亚男, 等. 代谢工程优化大肠杆菌高效合成L-苯丙氨酸[J]. 食品科学, 2021, 42(2): 114-120. |
| Men JX, Xiong B, Hao YN, et al. Metabolic engineering of Escherichia coli for efficient synthesis of L-phenylalanine[J]. Food Sci, 2021, 42(2): 114-120. | |
| [9] |
Liu SP, Xiao MR, Zhang L, et al. Production of l-phenylalanine from glucose by metabolic engineering of wild type Escherichia coli W3110[J]. Process Biochem, 2013, 48(3): 413-419.
doi: 10.1016/j.procbio.2013.02.016 URL |
| [10] |
Liu YJ, Li PP, Zhao KX, et al. Corynebacterium glutamicum contains 3-deoxy-D-arabino-heptulosonate 7-phosphate synthases that display novel biochemical features[J]. Appl Environ Microbiol, 2008, 74(17): 5497-5503.
doi: 10.1128/AEM.00262-08 URL |
| [11] |
Shu CH, Liao CC. Optimization of L-phenylalanine production of Corynebacterium glutamicum under product feedback inhibition by elevated oxygen transfer rate[J]. Biotechnol Bioeng, 2002, 77(2): 131-141.
doi: 10.1002/(ISSN)1097-0290 URL |
| [12] |
Dong C, Fontana J, Patel A, et al. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria[J]. Nat Commun, 2018, 9(1): 1-11.
doi: 10.1038/s41467-017-02088-w |
| [13] |
Lian JZ, HamediRad M, Hu SM, et al. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system[J]. Nat Commun, 2017, 8(1): 1-9.
doi: 10.1038/s41467-016-0009-6 |
| [14] |
Qi L, Larson M, Gilbert L, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression[J]. Cell, 2013, 152(5): 1173-1183.
doi: 10.1016/j.cell.2013.02.022 pmid: 23452860 |
| [15] |
Wang Y, Cheng HJ, Liu Y, et al. In-situ generation of large numbers of genetic combinations for metabolic reprogramming via CRISPR-guided base editing[J]. Nat Commun, 2021, 12: 678.
doi: 10.1038/s41467-021-21003-y pmid: 33514753 |
| [16] |
Heider SAE, Peters-Wendisch P, Wendisch VF. Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum[J]. BMC Microbiol, 2012, 12(1): 198.
doi: 10.1186/1471-2180-12-198 |
| [17] | 王猛, 王瑾. 一种小分子荧光传感器及其应用: CN114689552A[P]. 2022-07-01. |
| Meng W, Jin W. Small molecule fluorescence sensor and application thereof: CN114689552A[P]. 2022-07-01. | |
| [18] |
Liu X, Painter RE, Enesa K, et al. High-throughput screening of antibiotic-resistant bacteria in picodroplets[J]. Lab Chip, 2016, 16(9): 1636-1643.
doi: 10.1039/c6lc00180g pmid: 27033300 |
| [19] |
Mazutis L, Gilbert J, Ung WL, et al. Single-cell analysis and sorting using droplet-based microfluidics[J]. Nat Protoc, 2013, 8(5): 870-891.
doi: 10.1038/nprot.2013.046 pmid: 23558786 |
| [20] | 李俊维, 刘叶, 王钰, 等. 谷氨酸棒杆菌碱基编辑的条件优化[J]. 生物工程学报, 2020, 36(1): 143-151. |
| Li JW, Liu Y, Wang Y, et al. Optimization of base editing in Corynebacterium glutamicum[J]. Chin J Biotechnol, 2020, 36(1): 143-151. | |
| [21] |
Ruan YL, Zhu LJ, Li Q. Improving the electro-transformation efficiency of Corynebacterium glutamicum by weakening its cell wall and increasing the cytoplasmic membrane fluidity[J]. Biotechnol Lett, 2015, 37(12): 2445-2452.
doi: 10.1007/s10529-015-1934-x URL |
| [22] |
Schäfer A, Tauch A, Jäger W, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum[J]. Gene, 1994, 145(1): 69-73.
doi: 10.1016/0378-1119(94)90324-7 pmid: 8045426 |
| [23] |
Kostyuk AI, Demidovich AD, Kotova DA, et al. Circularly permuted fluorescent protein-based indicators: history, principles, and classification[J]. Int J Mol Sci, 2019, 20(17): 4200.
doi: 10.3390/ijms20174200 URL |
| [24] |
Chudakov DM, Matz MV, Lukyanov S, et al. Fluorescent proteins and their applications in imaging living cells and tissues[J]. Physiol Rev, 2010, 90(3): 1103-1163.
doi: 10.1152/physrev.00038.2009 pmid: 20664080 |
| [25] |
Zhang CZ, Zhang JL, Kang Z, et al. Enhanced production of l-phenylalanine in Corynebacterium glutamicum due to the introduction of Escherichia coli wild-type gene aroH[J]. J Ind Microbiol Biotechnol, 2013, 40(6): 643-651.
doi: 10.1007/s10295-013-1262-x URL |
| [26] |
Liu XZ, Niu H, Li Q, et al. Metabolic engineering for the production of l-phenylalanine in Escherichia coli[J]. 3 Biotech, 2019, 9(3): 85.
doi: 10.1007/s13205-019-1619-6 |
| [1] | 刘佳慧, 刘叶, 花尔并, 王猛. 谷氨酸棒杆菌中胞嘧啶碱基编辑工具的PAM拓展[J]. 生物技术通报, 2023, 39(9): 49-57. |
| [2] | 李雨真, 梅天秀, 李治文, 王淇, 李俊, 邹岳, 赵心清. 红酵母基因组和代谢工程改造研究进展[J]. 生物技术通报, 2023, 39(7): 67-79. |
| [3] | 李仁瀚, 张乐乐, 刘春立, 刘秀霞, 白仲虎, 杨艳坤, 李业. 基于紫色杆菌素生物合成途径的L-色氨酸生物传感器的构建[J]. 生物技术通报, 2023, 39(10): 80-92. |
| [4] | 刘金升, 陈振娅, 霍毅欣, 郭淑元. FACS技术在酶定向进化中的应用[J]. 生物技术通报, 2023, 39(10): 93-106. |
| [5] | 聂立斌, 易铃欣, 邓妍, 盛琦, 吴晓玉, 张斌. 途径工程改造谷氨酸棒杆菌产莽草酸[J]. 生物技术通报, 2022, 38(6): 93-102. |
| [6] | 张雪, 谭玉萌, 蒋海霞, 杨广宇. 基于单细胞超高通量筛选的α-1,2-岩藻糖基转移酶定向进化[J]. 生物技术通报, 2022, 38(1): 289-298. |
| [7] | 王鹏飞, 杨敏, 朱龙佼, 许文涛. 基于铂纳米团簇的生物传感研究进展[J]. 生物技术通报, 2021, 37(12): 235-242. |
| [8] | 赵颖, 王楠, 陆安祥, 冯晓元, 郭晓军, 栾云霞. 核酸适配体侧流层析分析技术在真菌毒素检测中的应用[J]. 生物技术通报, 2020, 36(8): 217-227. |
| [9] | 方顺燕, 宋丹, 刘艳萍, 徐文娟, 刘佳瑶, 韩向峙, 龙峰. 用于Escherichia coli O157∶H7直接快速检测的倏逝波荧光核酸适配体传感器研究[J]. 生物技术通报, 2020, 36(7): 228-234. |
| [10] | 叶健文, 陈江楠, 张旭, 吴赴清, 陈国强. 动态调控:一种高效的细胞工厂工程化代谢改造策略[J]. 生物技术通报, 2020, 36(6): 1-12. |
| [11] | 杨敏, 李舒婷, 杨文平, 李相阳, 许文涛. DNA/银纳米簇介导的功能核酸生物传感器研究进展[J]. 生物技术通报, 2020, 36(6): 245-254. |
| [12] | 柳苏月, 田晶晶, 田洪涛, 许文涛. 铽(III)离子及其复合物:从发光特性到传感应用[J]. 生物技术通报, 2020, 36(4): 192-207. |
| [13] | 孙雨阁, 李宸葳, 杜再慧, 许文涛. FEN1酶介导的功能核酸生物传感器的研究进展[J]. 生物技术通报, 2020, 36(4): 208-224. |
| [14] | 黄华媚, 白立宽, 刘叶, 李俊维, 王猛, 花尔并. BE3型胞嘧啶碱基编辑器在谷氨酸棒杆菌中的开发用[J]. 生物技术通报, 2020, 36(3): 95-101. |
| [15] | 聂志华, 朱蕾蕾. 生物素对发酵过程中MscCG外排L-谷氨酸的影响[J]. 生物技术通报, 2020, 36(10): 150-155. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||