








生物技术通报 ›› 2023, Vol. 39 ›› Issue (10): 1-16.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0243
• 综述与专论 • 下一篇
安昌1,2( ), 陆琳1, 沈梦千3, 陈盛圳3, 叶康卓1, 秦源1,2,3(
), 陆琳1, 沈梦千3, 陈盛圳3, 叶康卓1, 秦源1,2,3( ), 郑平1(
), 郑平1( )
)
收稿日期:2023-03-17
出版日期:2023-10-26
发布日期:2023-11-28
通讯作者:
郑平,女,副教授,研究方向:植物生殖发育及药用植物活性成分调控组学;E-mail: zhengping13@mails.ucas.ac.cn;作者简介:安昌,男,博士研究生,研究方向:中药资源分子鉴定及中药品质形成遗传机理;E-mail: ancher0928@163.com
基金资助:
AN Chang1,2( ), LU Lin1, SHEN Meng-qian3, CHEN Sheng-zhen3, YE Kang-zhuo1, QIN Yuan1,2,3(
), LU Lin1, SHEN Meng-qian3, CHEN Sheng-zhen3, YE Kang-zhuo1, QIN Yuan1,2,3( ), ZHENG Ping1(
), ZHENG Ping1( )
)
Received:2023-03-17
Published:2023-10-26
Online:2023-11-28
摘要:
碱性/螺旋-环-螺旋(bHLH)转录因子是植物中第二大转录因子家族,该家族广泛存在于各种植物的基因组中,并在植物生长发育、次生代谢、非生物逆境胁迫响应等方面发挥着重要的调控作用。本文全面综述了植物bHLH基因家族成员的结构特征、分类规则及其生物学功能的研究进展,重点梳理总结了bHLH在植物生长发育和非生物胁迫(干旱、低温、盐、重金属)中的应答和调控,以及在次生代谢产物生物合成及动态积累过程中的重要作用,可为深入研究bHLH在生长发育、植物抗逆及品质形成等方面的分子调控机制及种质资源的开发提供指导。同时,因bHLH广泛参与调控植物次生代谢产物的合成和积累,已成为分子生药学和中药生态农业研究的热点。为此,本文进一步总结了近年来研究较为透彻的两种药用植物(丹参Salvia Miltiorrhiza、黄花蒿Artemisia annua)中bHLH基因家族及其成员的研究进展,以期为药用植物bHLH基因家族的深入研究提供参考,并为药用植物的分子育种、拟境栽培等工作的开展以及中药生态农业的发展提供新思路。
安昌, 陆琳, 沈梦千, 陈盛圳, 叶康卓, 秦源, 郑平. 植物bHLH基因家族研究进展及在药用植物中的应用前景[J]. 生物技术通报, 2023, 39(10): 1-16.
AN Chang, LU Lin, SHEN Meng-qian, CHEN Sheng-zhen, YE Kang-zhuo, QIN Yuan, ZHENG Ping. Research Progress of bHLH Gene Family in Plants and Its Application Prospects in Medical Plants[J]. Biotechnology Bulletin, 2023, 39(10): 1-16.
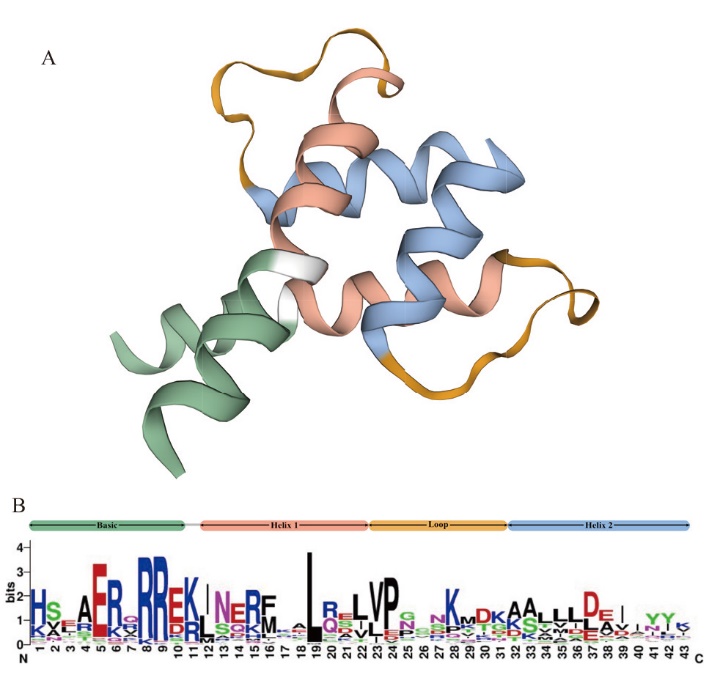
图1 黄花蒿bHLH家族成员结构域特征与分布 A:bHLH的三维蛋白质结构;B:WebLogo中生成的bHLH域的序列标识
Fig. 1 Domain characteristics and distribution of bHLH family members from A. annua A: Three-dimensional protein structure of bHLH. B: Sequence logo of the bHLH domain generated in WebLogo
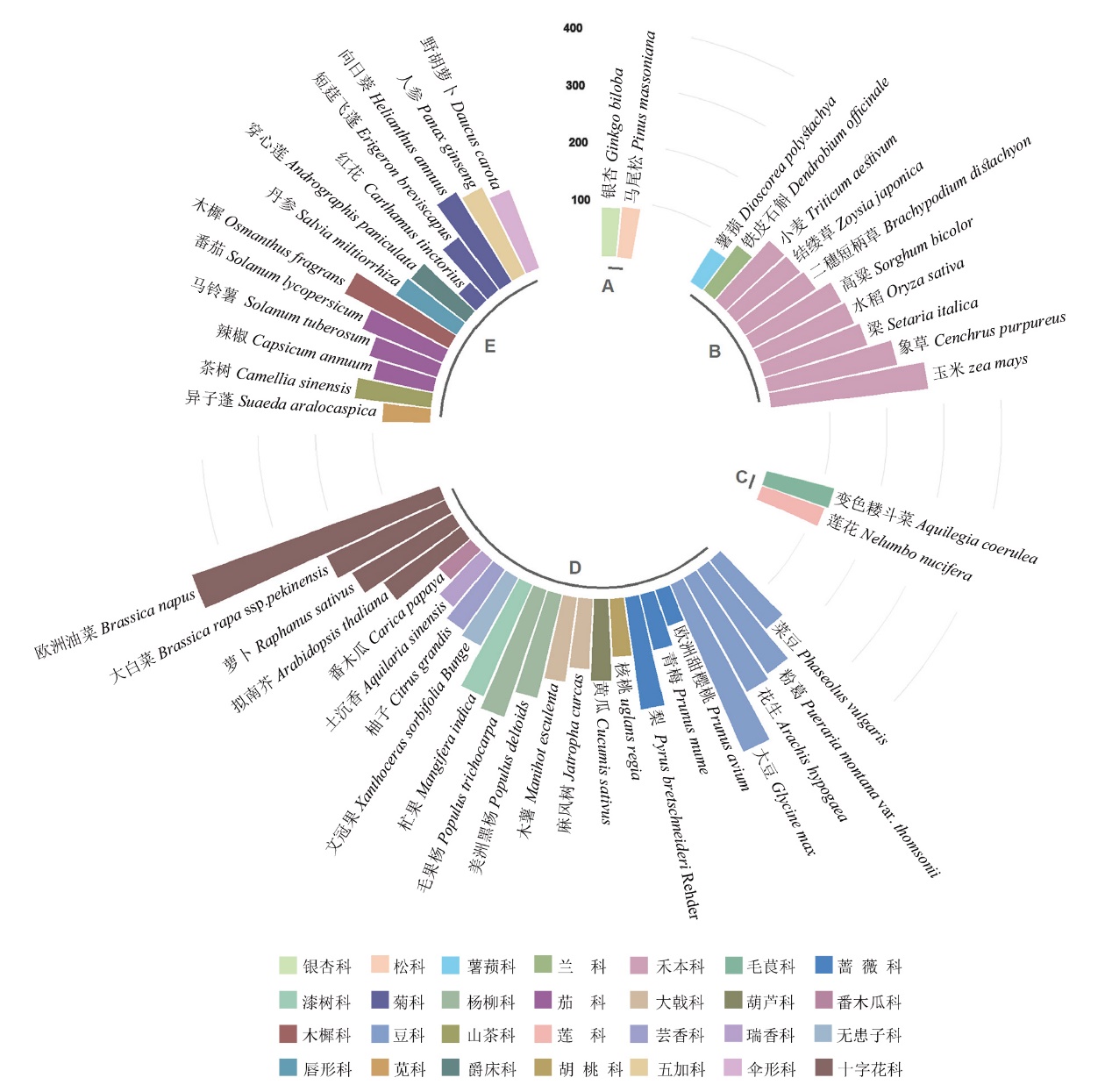
图2 部分种子植物中bHLH家族成员个数统计 A:裸子植物;B:单子叶植物;C:真双子叶植物基部群;D:核心真双子叶植物(超蔷薇类);E:核心真双子叶植物(超菊类)
Fig. 2 Statistics of the number of bHLH family members in vascular plants A: Gymnosperms. B: Monocots. C: Basal groups of eudicots. D: Core eudicots(superrosids). E: Core eudicots(superasterids)
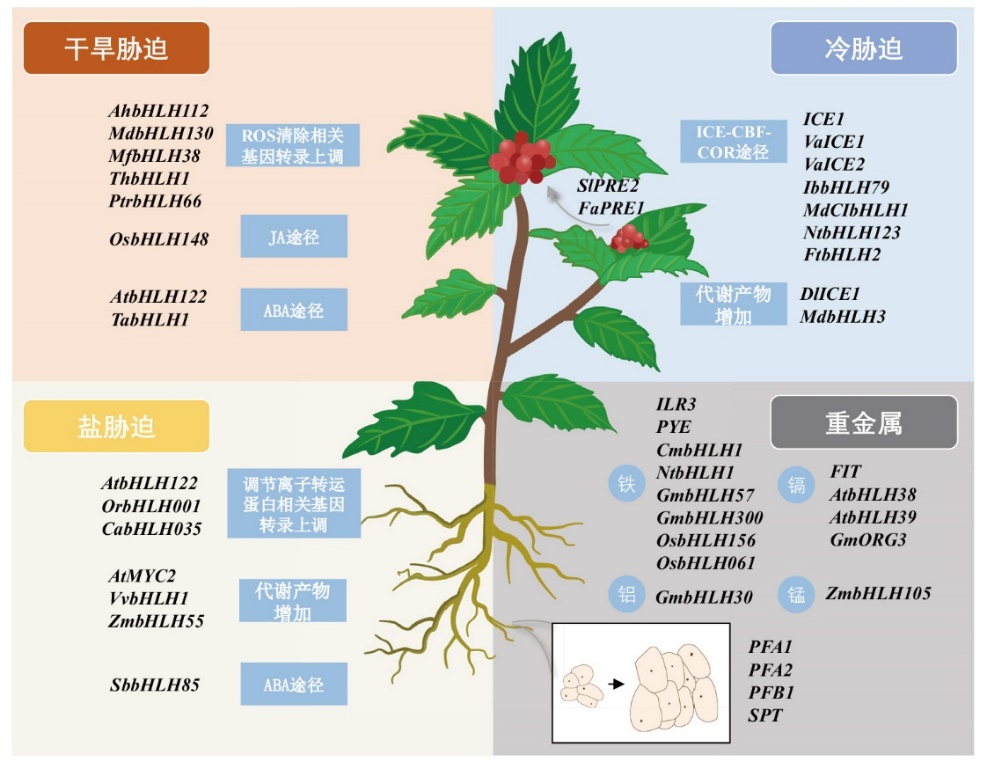
图3 bHLH家族成员参与植物生长发育和非生物胁迫过程的预测模型
Fig. 3 A predictive model for the involvement of bHLH family members in the growth and development of plants and abiotic stress processes
| [1] |
Rowe N, Speck T. Plant growth forms: an ecological and evolutionary perspective[J]. New Phytol, 2005, 166(1): 61-72.
pmid: 15760351 |
| [2] |
Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors[J]. Curr Opin Plant Biol, 2000, 3(5): 423-434.
pmid: 11019812 |
| [3] | 郭兰萍, 康传志, 周涛, 等. 中药生态农业最新进展及展望[J]. 中国中药杂志, 2021, 46(8): 1851-1857. |
| Guo LP, Kang CZ, Zhou T, et al. Ecological agriculture of Chinese materia medica: update and future perspectives[J]. China J Chin Mater Med, 2021, 46(8): 1851-1857. | |
| [4] | 郭兰萍, 周良云, 康传志, 等. 药用植物适应环境胁迫的策略及道地药材“拟境栽培”[J]. 中国中药杂志, 2020, 45(9): 1969-1974. |
| Guo LP, Zhou LY, Kang CZ, et al. Strategies for medicinal plants adapting environmental stress and “simulative habitat cultivation” of Dao-di herbs[J]. China J Chin Mater Med, 2020, 45(9): 1969-1974. | |
| [5] | 蒋待泉, 王红阳, 康传志, 等. 复合胁迫对药用植物次生代谢的影响及机制[J]. 中国中药杂志, 2020, 45(9): 2009-2016. |
| Jiang DQ, Wang HY, Kang CZ, et al. Influence and mechanism of stress combination on medicinal plants secondary metabolism[J]. China J Chin Mater Med, 2020, 45(9): 2009-2016. | |
| [6] | Sun W, Xu ZC, Song C, et al. Herbgenomics: decipher molecular genetics of medicinal plants[J]. Innovation(Camb), 2022, 3(6): 100322. |
| [7] |
Feller A, Machemer K, Braun EL, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors[J]. Plant J, 2011, 66(1): 94-116.
doi: 10.1111/tpj.2011.66.issue-1 URL |
| [8] | 鄢丹, 王伽伯, 李俊贤, 等. 论道地药材品质辨识及其与生态环境的相关性研究策略[J]. 中国中药杂志, 2012, 37(17): 2672-2675. |
| Yan D, Wang JB, Li JX, et al. Strategy for research on quality identification and ecological environment-related of Dao-di herb[J]. China J Chin Mater Med, 2012, 37(17): 2672-2675. | |
| [9] |
Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis[J]. Genome Res, 2001, 11(5): 754-770.
doi: 10.1101/gr.177001 pmid: 11337472 |
| [10] |
Jones S. An overview of the basic helix-loop-helix proteins[J]. Genome Biol, 2004, 5(6): 226.
doi: 10.1186/gb-2004-5-6-226 pmid: 15186484 |
| [11] |
Ludwig SR, Habera LF, Dellaporta SL, et al. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region[J]. Proc Natl Acad Sci USA, 1989, 86(18): 7092-7096.
doi: 10.1073/pnas.86.18.7092 pmid: 2674946 |
| [12] |
Zhang TT, Lyu W, Zhang HS, et al. Genome-wide analysis of the basic helix-loop-helix(bHLH)transcription factor family in maize[J]. BMC Plant Biol, 2018, 18(1): 235.
doi: 10.1186/s12870-018-1441-z |
| [13] |
Skinner MK, Rawls A, Wilson-Rawls J, et al. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature[J]. Differentiation, 2010, 80(1): 1-8.
doi: 10.1016/j.diff.2010.02.003 pmid: 20219281 |
| [14] |
Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family[J]. Plant Cell, 2003, 15(8): 1749-1770.
doi: 10.1105/tpc.013839 pmid: 12897250 |
| [15] |
Roig-Villanova I, Bou-Torrent J, Galstyan A, et al. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins[J]. EMBO J, 2007, 26(22): 4756-4767.
doi: 10.1038/sj.emboj.7601890 pmid: 17948056 |
| [16] |
Medina-Puche L, Martínez-Rivas FJ, Molina-Hidalgo FJ, et al. An atypical HLH transcriptional regulator plays a novel and important role in strawberry ripened receptacle[J]. BMC Plant Biol, 2019, 19(1): 586.
doi: 10.1186/s12870-019-2092-4 pmid: 31881835 |
| [17] |
Carretero-Paulet L, Galstyan A, Roig-Villanova I, et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae[J]. Plant Physiol, 2010, 153(3): 1398-1412.
doi: 10.1104/pp.110.153593 pmid: 20472752 |
| [18] | 位志坤, 许自成, 贾国涛. bHLH转录因子家族调控植物非生物逆境胁迫响应的研究进展[J/OL]. 分子植物育种, 2022. https://kns.cnki.net/kcms/detail/46.1068.S.20220615.0859.002.html. |
| Wei ZK, Xu ZC, Jia GT, et al. BHLH transcription factor family regulates abiotic stress response in plants research progress[J/OL]. Mol Breed, 2022. https://kns.cnki.net/kcms/detail/46.1068.S.20220615.0859.002.html. | |
| [19] |
Hong YQ, Ahmad N, Tian YY, et al. Genome-wide identification, expression analysis, and subcellular localization of Carthamus tinctorius bHLH transcription factors[J]. Int J Mol Sci, 2019, 20(12): 3044.
doi: 10.3390/ijms20123044 URL |
| [20] |
Shen W, Cui X, Li H, et al. Genome-wide identification and analyses of bHLH family genes in Brassica napus[J]. Can J Plant Sci, 2019, 99(5): 589-598.
doi: 10.1139/cjps-2018-0230 |
| [21] |
Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants[J]. Mol Biol Evol, 2010, 27(4): 862-874.
doi: 10.1093/molbev/msp288 pmid: 19942615 |
| [22] |
Gao F, Robe K, Gaymard F, et al. The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors?[J]. Front Plant Sci, 2019, 10: 6.
doi: 10.3389/fpls.2019.00006 pmid: 30713541 |
| [23] |
Gao C, Sun JL, Wang CQ, et al. Genome-wide analysis of basic/helix-loop-helix gene family in peanut and assessment of its roles in pod development[J]. PLoS One, 2017, 12(7): e0181843.
doi: 10.1371/journal.pone.0181843 URL |
| [24] |
Mao TY, Liu YY, Zhu HH, et al. Genome-wide analyses of the bHLH gene family reveals structural and functional characteristics in the aquatic plant Nelumbo nucifera[J]. PeerJ, 2019, 7: e7153.
doi: 10.7717/peerj.7153 URL |
| [25] |
Chu Y, Xiao SM, Su H, et al. Genome-wide characterization and analysis of bHLH transcription factors in Panax ginseng[J]. Acta Pharm Sin B, 2018, 8(4): 666-677.
doi: 10.1016/j.apsb.2018.04.004 URL |
| [26] |
Mauxion JP, Chevalier C, Gonzalez N. Complex cellular and molecular events determining fruit size[J]. Trends Plant Sci, 2021, 26(10): 1023-1038.
doi: 10.1016/j.tplants.2021.05.008 URL |
| [27] |
Zhu ZG, Chen GP, Guo XH, et al. Overexpression of SlPRE2, an atypical bHLH transcription factor, affects plant morphology and fruit pigment accumulation in tomato[J]. Sci Rep, 2017, 7(1): 5786.
doi: 10.1038/s41598-017-04092-y |
| [28] |
Zhu ZG, Liang HL, Chen GP, et al. The bHLH transcription factor SlPRE2 regulates tomato fruit development and modulates plant response to gibberellin[J]. Plant Cell Rep, 2019, 38(9): 1053-1064.
doi: 10.1007/s00299-019-02425-x pmid: 31123809 |
| [29] |
Tan C, Qiao HL, Ma M, et al. Genome-wide identification and characterization of melon bHLH transcription factors in regulation of fruit development[J]. Plants, 2021, 10(12): 2721.
doi: 10.3390/plants10122721 URL |
| [30] |
Yu JQ, Gu KD, Sun CH, et al. The apple bHLH transcription factor MdbHLH3 functions in determining the fruit carbohydrates and malate[J]. Plant Biotechnol J, 2021, 19(2): 285-299.
doi: 10.1111/pbi.v19.2 URL |
| [31] |
Zhang Y, Mitsuda N, Yoshizumi T, et al. Two types of bHLH transcription factor determine the competence of the pericycle for lateral root initiation[J]. Nat Plants, 2021, 7(5): 633-643.
doi: 10.1038/s41477-021-00919-9 pmid: 34007039 |
| [32] |
Groszmann M, Paicu T, Smyth DR. Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis[J]. Plant J, 2008, 55(1): 40-52.
doi: 10.1111/tpj.2008.55.issue-1 URL |
| [33] |
Ichihashi Y, Horiguchi G, Gleissberg S, et al. The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana[J]. Plant Cell Physiol, 2010, 51(2): 252-261.
doi: 10.1093/pcp/pcp184 pmid: 20040585 |
| [34] |
Groszmann M, Paicu T, Alvarez JP, et al. SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development[J]. Plant J, 2011, 68(5): 816-829.
doi: 10.1111/tpj.2011.68.issue-5 URL |
| [35] |
Makkena S, Lamb RS. The bHLH transcription factor SPATULA is a key regulator of organ size in Arabidopsis thaliana[J]. Plant Signal Behav, 2013, 8(5): e24140.
doi: 10.4161/psb.24140 URL |
| [36] |
Makkena S, Lamb RS. The bHLH transcription factor SPATULA regulates root growth by controlling the size of the root meristem[J]. BMC Plant Biol, 2013, 13: 1.
doi: 10.1186/1471-2229-13-1 URL |
| [37] |
Li HM, Sun JQ, Xu YX, et al. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis[J]. Plant Mol Biol, 2007, 65(5): 655-665.
doi: 10.1007/s11103-007-9230-3 URL |
| [38] |
Liu WW, Tai HH, Li SS, et al. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism[J]. New Phytol, 2014, 201(4): 1192-1204.
doi: 10.1111/nph.2014.201.issue-4 URL |
| [39] |
Yang TR, Yao SF, Hao L, et al. Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway[J]. Plant Cell Rep, 2016, 35(11): 2309-2323.
doi: 10.1007/s00299-016-2036-5 URL |
| [40] |
Seo JS, Joo J, Kim MJ, et al. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice[J]. Plant J, 2011, 65(6): 907-921.
doi: 10.1111/tpj.2011.65.issue-6 URL |
| [41] |
Castilhos G, Lazzarotto F, Spagnolo-Fonini L, et al. Possible roles of basic helix-loop-helix transcription factors in adaptation to drought[J]. Plant Sci, 2014, 223: 1-7.
doi: 10.1016/j.plantsci.2014.02.010 pmid: 24767109 |
| [42] |
Li CJ, Yan CX, Sun QX, et al. The bHLH transcription factor AhbHLH112 improves the drought tolerance of peanut[J]. BMC Plant Biol, 2021, 21(1): 540.
doi: 10.1186/s12870-021-03318-6 pmid: 34784902 |
| [43] |
Zhao Q, Fan ZH, Qiu LN, et al. MdbHLH130, an apple bHLH transcription factor, confers water stress resistance by regulating stomatal closure and ROS homeostasis in transgenic tobacco[J]. Front Plant Sci, 2020, 11: 543696.
doi: 10.3389/fpls.2020.543696 URL |
| [44] |
Dong Y, Wang CP, Han X, et al. A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis[J]. Biochem Biophys Res Commun, 2014, 450(1): 453-458.
doi: 10.1016/j.bbrc.2014.05.139 URL |
| [45] |
Qiu JR, Huang Z, Xiang XY, et al. MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis[J]. BMC Plant Biol, 2020, 20(1): 542.
doi: 10.1186/s12870-020-02732-6 |
| [46] | Ji XY, Nie XG, Liu YJ, et al. A bHLH gene from Tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation[J]. Tree Physiol, 2016, 36(2): 193-207. |
| [72] |
Wang WJ, Shinwari KI, Zhang H, et al. The bHLH transcription factor OsbHLH057 regulates iron homeostasis in rice[J]. Int J Mol Sci, 2022, 23(23): 14869.
doi: 10.3390/ijms232314869 URL |
| [73] |
Wang WJ, Ye J, Xu H, et al. OsbHLH061 links topless/topless-related repressor proteins with positive regulator of iron homeostasis 1 to maintain iron homeostasis in rice[J]. New Phytol, 2022, 234(5): 1753-1769.
doi: 10.1111/nph.18096 pmid: 35288933 |
| [74] |
Cui YC, Xu GY, Wang ML, et al. Expression of OsMSR3 in Arabidopsis enhances tolerance to cadmium stress[J]. Plant Cell Tiss Organ Cult, 2013, 113(2): 331-340.
doi: 10.1007/s11240-012-0275-x URL |
| [75] |
Xu ZL, Liu XQ, He XL, et al. The soybean basic hlix-loop-hlix transcription factor ORG3-like enhances cadmium tolerance via increased iron and reduced cadmium uptake and transport from roots to shoots[J]. Front Plant Sci, 2017, 8: 1098.
doi: 10.3389/fpls.2017.01098 URL |
| [76] | 宋倩. 铝胁迫下大豆GmbHLH30转录因子调控相关基因的分离与鉴定[D]. 昆明: 昆明理工大学, 2014. |
| Song Q. Isolation and characterization of relate genes regulated by soybean GmbHLH30 transcription factor under Al stress[D]. Kunming: Kunming University of Science and Technology, 2014. | |
| [77] |
Sun KL, Wang HY, Xia ZL. The maize bHLH transcription factor bHLH105 confers manganese tolerance in transgenic tobacco[J]. Plant Sci, 2019, 280: 97-109.
doi: S0168-9452(18)31005-7 pmid: 30824033 |
| [78] |
Hichri I, Barrieu F, Bogs J, et al. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway[J]. J Exp Bot, 2011, 62(8): 2465-2483.
doi: 10.1093/jxb/erq442 pmid: 21278228 |
| [79] |
McClean PE, Lee RA, Howe K, et al. The common bean V gene encodes flavonoid 3'5' hydroxylase: a major mutational target for flavonoid diversity in angiosperms[J]. Front Plant Sci, 2022, 13: 869582.
doi: 10.3389/fpls.2022.869582 URL |
| [80] |
Baudry A, Caboche M, Lepiniec L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana[J]. Plant J, 2006, 46(5): 768-779.
doi: 10.1111/tpj.2006.46.issue-5 URL |
| [47] |
Liang BB, Wan SG, Ma QL, et al. A novel bHLH transcription factor PtrbHLH66 from trifoliate orange positively regulates plant drought tolerance by mediating root growth and ROS scavenging[J]. Int J Mol Sci, 2022, 23(23): 15053.
doi: 10.3390/ijms232315053 URL |
| [48] |
Liu H, Yang Y, Liu DD, et al. Transcription factor TabHLH49 positively regulates dehydrin WZY2 gene expression and enhances drought stress tolerance in wheat[J]. BMC Plant Biol, 2020, 20(1): 259.
doi: 10.1186/s12870-020-02474-5 |
| [49] |
Gu XY, Gao SX, Li J, et al. The bHLH transcription factor regulated gene OsWIH2 is a positive regulator of drought tolerance in rice[J]. Plant Physiol Biochem, 2021, 169: 269-279.
doi: 10.1016/j.plaphy.2021.11.031 URL |
| [50] |
Kim J, Kim HY. Functional analysis of a calcium-binding transcription factor involved in plant salt stress signaling[J]. FEBS Lett, 2006, 580(22): 5251-5256.
pmid: 16962584 |
| [51] |
Krishnamurthy P, Vishal B, Khoo K, et al. Expression of AoN-HX1 increases salt tolerance of rice and Arabidopsis, and bHLH transcription factors regulate AtNHX1 and AtNHX6 in Arabidop-sis[J]. Plant Cell Rep, 2019, 38(10): 1299-1315.
doi: 10.1007/s00299-019-02450-w pmid: 31350571 |
| [52] |
Verma D, Jalmi SK, Bhagat PK, et al. A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis[J]. FEBS J, 2020, 287(12): 2560-2576.
doi: 10.1111/febs.v287.12 URL |
| [53] |
Zhang HF, Guo JB, Chen XQ, et al. Pepper bHLH transcription factor CabHLH035 contributes to salt tolerance by modulating ion homeostasis and proline biosynthesis[J]. Hortic Res, 2022, 9: uhac203.
doi: 10.1093/hr/uhac203 URL |
| [54] |
Wang FB, Zhu H, Chen DH, et al. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana[J]. Plant Cell Tiss Organ Cult, 2016, 125(2): 387-398.
doi: 10.1007/s11240-016-0953-1 URL |
| [55] |
Yu CM, Yan M, Dong HZ, et al. Maize bHLH55 functions positively in salt tolerance through modulation of AsA biosynthesis by directly regulating GDP-mannose pathway genes[J]. Plant Sci, 2021, 302: 110676.
doi: 10.1016/j.plantsci.2020.110676 URL |
| [56] |
Song YS, Li JL, Sui Y, et al. The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis[J]. Plant Mol Biol, 2020, 102(6): 603-614.
doi: 10.1007/s11103-020-00966-4 pmid: 32052233 |
| [57] |
Zhang KY, Zhang ZP, Lu F, et al. Bulked segregant analysis-sequencing identification of candidate genes for salt tolerance at the seedling stage of sorghum(Sorghum bicolor)[J]. Plant Breed, 2022, 141(3): 366-378.
doi: 10.1111/pbr.v141.3 URL |
| [58] |
Song YS, Li SM, Sui Y, et al. SbbHLH85, a bHLH member, modulates resilience to salt stress by regulating root hair growth in sorghum[J]. Theor Appl Genet, 2022, 135(1): 201-216.
doi: 10.1007/s00122-021-03960-6 |
| [59] |
Chinnusamy V, Ohta M, Kanrar S, et al. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis[J]. Genes Dev, 2003, 17(8): 1043-1054.
doi: 10.1101/gad.1077503 URL |
| [60] |
Xu WR, Jiao YT, Li RM, et al. Chinese wild-growing Vitis amurensis ICE1 and ICE2 encode MYC-type bHLH transcription activators that regulate cold tolerance in Arabidopsis[J]. PLoS One, 2014, 9(7): e102303.
doi: 10.1371/journal.pone.0102303 URL |
| [61] |
Jin R, Kim HS, Yu T, et al. Identification and function analysis of bHLH genes in response to cold stress in sweetpotato[J]. Plant Physiol Biochem, 2021, 169: 224-235.
doi: 10.1016/j.plaphy.2021.11.027 URL |
| [62] |
Verma RK, Kumar VVS, Yadav SK, et al. Overexpression of Arabidopsis ICE1 enhances yield and multiple abiotic stress tolerance in indica rice[J]. Plant Signal Behav, 2020, 15(11): 1814547.
doi: 10.1080/15592324.2020.1814547 URL |
| [63] |
Yao PF, Sun ZX, Li CL, et al. Overexpression of Fagopyrum tataricum FtbHLH2 enhances tolerance to cold stress in transgenic Arabidopsis[J]. Plant Physiol Biochem, 2018, 125: 85-94.
doi: 10.1016/j.plaphy.2018.01.028 URL |
| [64] |
Yang XY, Wang R, Hu QL, et al. DlICE1, a stress-responsive gene from Dimocarpus longan, enhances cold tolerance in transgenic Arabidopsis[J]. Plant Physiol Biochem, 2019, 142: 490-499.
doi: 10.1016/j.plaphy.2019.08.007 URL |
| [65] |
Thao NP, Khan MI, Thu NBA, et al. Role of ethylene and its cross talk with other signaling molecules in plant responses to heavy metal stress[J]. Plant Physiol, 2015, 169(1): 73-84.
doi: 10.1104/pp.15.00663 pmid: 26246451 |
| [66] |
Kim SA, LaCroix IS, Gerber SA, et al. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI[J]. Proc Natl Acad Sci USA, 2019, 116(50): 24933-24942.
doi: 10.1073/pnas.1916892116 URL |
| [81] |
Deng J, Li JJ, Su MY, et al. A bHLH gene NnTT8 of Nelumbo nucifera regulates anthocyanin biosynthesis[J]. Plant Physiol Biochem, 2021, 158: 518-523.
doi: 10.1016/j.plaphy.2020.11.038 URL |
| [82] |
Jia N, Wang JJ, Liu JM, et al. DcTT8, a bHLH transcription factor, regulates anthocyanin biosynthesis in Dendrobium candidum[J]. Plant Physiol Biochem, 2021, 162: 603-612.
doi: 10.1016/j.plaphy.2021.03.006 URL |
| [83] |
Qi Y, Zhou L, Han LL, et al. PsbHLH1, a novel transcription factor involved in regulating anthocyanin biosynthesis in tree peony(Paeonia suffruticosa)[J]. Plant Physiol Biochem, 2020, 154: 396-408.
doi: 10.1016/j.plaphy.2020.06.015 URL |
| [84] |
Li H, Yang Z, Zeng QW, et al. Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits[J]. Hortic Res, 2020, 7(1): 83.
doi: 10.1038/s41438-020-0302-8 |
| [85] |
Zhao R, Song XX, Yang N, et al. Expression of the subgroup IIIf bHLH transcription factor CpbHLH1 from Chimonanthus praecox(L.) in transgenic model plants inhibits anthocyanin accumulation[J]. Plant Cell Rep, 2020, 39(7): 891-907.
doi: 10.1007/s00299-020-02537-9 |
| [86] | Zhang JH, Lyu HZ, Liu WJ, et al. bHLH transcription factor SmbHLH92 negatively regulates biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza[J]. Chin Herb Med, 2020, 12(3): 237-246. |
| [87] |
Xing BC, Yang DF, Yu HZ, et al. Overexpression of SmbHLH10 enhances tanshinones biosynthesis in Salvia miltiorrhiza hairy roots[J]. Plant Sci, 2018, 276: 229-238.
doi: 10.1016/j.plantsci.2018.07.016 URL |
| [88] |
Mertens J, Pollier J, Vanden Bossche R, et al. The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula[J]. Plant Physiol, 2016, 170(1): 194-210.
doi: 10.1104/pp.15.01645 pmid: 26589673 |
| [89] |
De Boer K, Tilleman S, Pauwels L, et al. apetala2/ethylene response factor and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis[J]. Plant J, 2011, 66(6): 1053-1065.
doi: 10.1111/tpj.2011.66.issue-6 URL |
| [90] |
Xiao L, Huang D, Wu ZD, et al. Genome-wide identification of the bHLH transcription factor family and analysis of bHLH genes related to puerarin biosynthesis in Pueraria lobata var. thomsonii(Benth.)[J]. Plant Gene, 2023, 33: 100390.
doi: 10.1016/j.plgene.2022.100390 URL |
| [67] |
Gao F, Robe K, Bettembourg M, et al. The transcription factor bHLH121 interacts with bHLH105(ILR3)and its closest homologs to regulate iron homeostasis in Arabidopsis[J]. Plant Cell, 2020, 32(2): 508-524.
doi: 10.1105/tpc.19.00541 URL |
| [68] | Akmakjian GZ, Riaz N, Guerinot ML. Photoprotection during iron deficiency is mediated by the bHLH transcription factors PYE and ILR3[J]. Proc Natl Acad Sci USA, 2021, 118(40): e2024918118. |
| [69] |
Li YY, Sui XY, Yang JS, et al. A novel bHLH transcription factor, NtbHLH1, modulates iron homeostasis in tobacco(Nicotiana tabacum L.)[J]. Biochem Biophys Res Commun, 2020, 522(1): 233-239.
doi: 10.1016/j.bbrc.2019.11.063 URL |
| [70] |
Li L, Gao WW, Peng Q, et al. Two soybean bHLH factors regulate response to iron deficiency[J]. J Integr Plant Biol, 2018, 60(7): 608-622.
doi: 10.1111/jipb.12651 |
| [71] |
Wang SD, Li L, Ying YH, et al. A transcription factor OsbHLH156 regulates Strategy II iron acquisition through localising IRO2 to the nucleus in rice[J]. New Phytol, 2020, 225(3): 1247-1260.
doi: 10.1111/nph.16232 pmid: 31574173 |
| [91] |
Sánchez-Pérez R, Pavan S, Mazzeo R, et al. Mutation of a bHLH transcription factor allowed almond domestication[J]. Science, 2019, 364(6445): 1095-1098.
doi: 10.1126/science.aav8197 pmid: 31197015 |
| [92] |
Zheng H, Fu XQ, Shao J, et al. Transcriptional regulatory network of high-value active ingredients in medicinal plants[J]. Trends Plant Sci, 2023, 28(4): 429-446.
doi: 10.1016/j.tplants.2022.12.007 pmid: 36621413 |
| [93] |
Zhang X, Luo HM, Xu ZC, et al. Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza[J]. Sci Rep, 2015, 5: 11244.
doi: 10.1038/srep11244 pmid: 26174967 |
| [94] | 李林. 基于茉莉素信号介导的丹参转录因子SmbHLH59、SmMYB97和SmWRKY14的功能研究[D]. 西安: 陕西师范大学, 2021. |
| Li L. Functional study on transcription factors SmbHLH59, SmMYB97 and SmWRKY14 of Salvia Miltiorrhiza based on Jasmine signal[D]. Xi'an: Shaanxi Normal University, 2021. | |
| [95] |
Wang H, Li SY, Li YA, et al. MED25 connects enhancer-promoter looping and MYC2-dependent activation of jasmonate signalling[J]. Nat Plants, 2019, 5(6): 616-625.
doi: 10.1038/s41477-019-0441-9 pmid: 31182849 |
| [96] |
Liu SC, Wang Y, Shi M, et al. SmbHLH60 and SmMYC2 antagonistically regulate phenolic acids and anthocyanins biosynthesis in Salvia miltiorrhiza[J]. J Adv Res, 2022, 42: 205-219.
doi: 10.1016/j.jare.2022.02.005 URL |
| [97] | 方庆. 基于SmbHLH124转录因子的丹参次生代谢工程研究[D]. 成都: 电子科技大学, 2021. |
| Fang Q. Metabolic engineering with transcription factor SmbHLH124 in Salvia miltiorrhiza[D]. Chengdu: University of Electronic Science and Technology of China, 2021. | |
| [98] | 甘雨, 吴端, 张栋, 等. 黄花蒿bHLH转录因子基因家族鉴定及光调控分析[J]. 中国现代中药, 2021, 23(3): 441-452. |
| Gan Y, Wu D, Zhang D, et al. Identification and light regulation analysis of bHLH transcription factor gene family in Artemisia annua[J]. Mod Chin Med, 2021, 23(3): 441-452. | |
| [99] |
Kayani SI, Shen Q, Ma YN, et al. The YABBY family transcription factor AaYABBY5 directly targets cytochrome P450 monooxygenase(CYP71AV1)and double-bond reductase 2(DBR2)involved in artemisinin biosynthesis in Artemisia annua[J]. Front Plant Sci, 2019, 10: 1084.
doi: 10.3389/fpls.2019.01084 URL |
| [100] |
Liao BS, Shen XF, Xiang L, et al. Allele-aware chromosome-level genome assembly of Artemisia annua reveals the correlation between ADS expansion and artemisinin yield[J]. Mol Plant, 2022, 15(8): 1310-1328.
doi: 10.1016/j.molp.2022.05.013 URL |
| [101] |
Gordân R, Shen N, Dror I, et al. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape[J]. Cell Rep, 2013, 3(4): 1093-1104.
doi: 10.1016/j.celrep.2013.03.014 pmid: 23562153 |
| [102] |
Le Dréau G, Escalona R, Fueyo R, et al. E proteins sharpen neurogenesis by modulating proneural bHLH transcription factors'activity in an E-box-dependent manner[J]. eLife, 2018, 7: e37267.
doi: 10.7554/eLife.37267 URL |
| [103] |
Shi P, Fu XQ, Shen Q, et al. The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in Artemisia annua[J]. New Phytol, 2018, 217(1): 261-276.
doi: 10.1111/nph.14789 pmid: 28940606 |
| [104] |
Ji YP, Xiao JW, Shen YL, et al. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua[J]. Plant Cell Physiol, 2014, 55(9): 1592-1604.
doi: 10.1093/pcp/pcu090 URL |
| [105] |
Zhang QZ, Wu NY, Jian DQ, et al. Overexpression of AaPIF3 promotes artemisinin production in Artemisia annua[J]. Ind Crops Prod, 2019, 138: 111476.
doi: 10.1016/j.indcrop.2019.111476 URL |
| [106] |
Xiang LE, Jian DQ, Zhang FY, et al. The cold-induced transcription factor bHLH112 promotes artemisinin biosynthesis indirectly via ERF1 in Artemisia annua[J]. J Exp Bot, 2019, 70(18): 4835-4848.
doi: 10.1093/jxb/erz220 pmid: 31087059 |
| [107] |
Bredow M, Vanderbeld B, Walker VK. Ice-binding proteins confer freezing tolerance in transgenic Arabidopsis thaliana[J]. Plant Biotechnol J, 2017, 15(1): 68-81.
doi: 10.1111/pbi.2017.15.issue-1 URL |
| [108] |
Shen Q, Huang HY, Xie LH, et al. Basic helix-loop-helix transcription factors AabHLH2 and AabHLH3 function antagonistically with AaMYC2 and are negative regulators in artemisinin biosynthesis[J]. Front Plant Sci, 2022, 13: 885622.
doi: 10.3389/fpls.2022.885622 URL |
| [109] | 周琪. AabHLH106调控青蒿非青蒿素倍半萜生物合成的功能研究[D]. 重庆: 西南大学, 2022. |
| Zhou Q. Functional characterization of AabHLH106 in the regulation on non-artemisinin sesquiterpenes biosynthesis in Artemisia annua[D]. Chongqing: Southwest University, 2022. | |
| [110] |
Heim MA, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity[J]. Mol Biol Evol, 2003, 20(5): 735-747.
doi: 10.1093/molbev/msg088 pmid: 12679534 |
| [111] |
Lamb P, McKnight SL. Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization[J]. Trends Biochem Sci, 1991, 16(11): 417-422.
pmid: 1776171 |
| [112] |
Amoutzias GD, Robertson DL, Van de Peer Y, et al. Choose your partners: dimerization in eukaryotic transcription factors[J]. Trends Biochem Sci, 2008, 33(5): 220-229.
doi: 10.1016/j.tibs.2008.02.002 pmid: 18406148 |
| [113] |
Ben Rejeb K, Abdelly C, Savouré A. How reactive oxygen species and proline face stress together[J]. Plant Physiol Biochem, 2014, 80: 278-284.
doi: 10.1016/j.plaphy.2014.04.007 URL |
| [114] |
Mittler R. Abiotic stress, the field environment and stress combination[J]. Trends Plant Sci, 2006, 11(1): 15-19.
doi: 10.1016/j.tplants.2005.11.002 pmid: 16359910 |
| [115] |
Zhu L, Zhao MZ, Chen MY, et al. The bHLH gene family and its response to saline stress in Jilin ginseng, Panax ginseng C.A. Meyer[J]. Mol Genet Genomics, 2020, 295(4): 877-890.
doi: 10.1007/s00438-020-01658-w |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [3] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [4] | 胡海琳, 徐黎, 李晓旭, 王晨璨, 梅曼, 丁文静, 赵媛媛. 小肽激素调控植物生长发育及逆境生理研究进展[J]. 生物技术通报, 2023, 39(7): 13-25. |
| [5] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [6] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [7] | 李苑虹, 郭昱昊, 曹燕, 祝振洲, 王飞飞. 外源植物激素调控微藻生长及目标产物积累研究进展[J]. 生物技术通报, 2023, 39(6): 61-72. |
| [8] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [9] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [10] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [11] | 薛皦, 朱庆锋, 冯彦钊, 陈沛, 刘文华, 张爱霞, 刘勤坚, 张琪, 于洋. 植物基因上游开放阅读框的研究进展[J]. 生物技术通报, 2023, 39(4): 157-165. |
| [12] | 魏明, 王欣玉, 伍国强, 赵萌. NAD依赖型去乙酰化酶SRT在植物表观遗传调控中的作用[J]. 生物技术通报, 2023, 39(4): 59-70. |
| [13] | 桑田, 王鹏程. 植物SUMO化修饰研究进展[J]. 生物技术通报, 2023, 39(3): 1-12. |
| [14] | 杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142. |
| [15] | 郑敏敏, 柳洁, 赵清. 药用植物黄芩的生物学研究进展及展望[J]. 生物技术通报, 2023, 39(2): 10-23. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||