








生物技术通报 ›› 2024, Vol. 40 ›› Issue (2): 120-129.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0697
陈凯凌1( ), 武涛1,2, 徐逸群1,2, 高佳1,2, 张美俊1,2, 李欣1(
), 武涛1,2, 徐逸群1,2, 高佳1,2, 张美俊1,2, 李欣1( ), 贾举庆1,2(
), 贾举庆1,2( )
)
收稿日期:2023-07-18
出版日期:2024-02-26
发布日期:2024-03-13
通讯作者:
李欣,男,硕士,副研究员,研究方向:作物遗传与育种;E-mail: lixin@sxau.edu.cn;作者简介:陈凯凌,男,硕士研究生,研究方向:作物遗传与育种;E-mail: ckl1228@qq.com
基金资助:
CHEN Kai-ling1( ), WU Tao1,2, XU Yi-qun1,2, GAO Jia1,2, ZHANG Mei-jun1,2, LI Xin1(
), WU Tao1,2, XU Yi-qun1,2, GAO Jia1,2, ZHANG Mei-jun1,2, LI Xin1( ), JIA Ju-qing1,2(
), JIA Ju-qing1,2( )
)
Received:2023-07-18
Published:2024-02-26
Online:2024-03-13
摘要:
【目的】了解燕麦基因组SSR位点分布情况并开发燕麦全基因组SSR标记,为燕麦皮裸基因克隆、遗传图谱构建、群体多样性分析等提供科学的参考依据。【方法】利用已公布的“三分三”裸燕麦参考基因组,通过TBtools软件对燕麦全基因组的SSR位点进行检索并利用EXCEL和SPSS软件对检索数据分析其分布特征。根据检索结果设计引物,在构建的F2遗传群体中小规模进行非变性聚丙烯酰胺凝胶电泳验证,并克隆测序其有效性与真实性。【结果】在燕麦基因组中,SSR数量众多,类型丰富。全基因组共检索到828 138个SSR位点,SSR平均密度为78.16个/Mb,平均距离为12.90 kb。单、双、三核苷酸重复类型占主导优势。基元重复次数主要集中在5次和6次,A/T、AG/CT和AAC/GTT重复基元为优势基元。燕麦基因组SSR位点呈现中度多态性,长度变化范围为11-1 587 bp。并由此开发设计52对多态性引物在试验材料中进行扩增验证,在构建的遗传群体中均具有多态性。【结论】在“三分三”裸燕麦参考基因组中开发SSR引物有效可行,可以用于后续进一步试验研究。
陈凯凌, 武涛, 徐逸群, 高佳, 张美俊, 李欣, 贾举庆. 燕麦全基因组SSR位点鉴定及多态性标记开发[J]. 生物技术通报, 2024, 40(2): 120-129.
CHEN Kai-ling, WU Tao, XU Yi-qun, GAO Jia, ZHANG Mei-jun, LI Xin, JIA Ju-qing. Identification of SSR Loci and Development of Polymorphic Markers in Whole Genome of Oat[J]. Biotechnology Bulletin, 2024, 40(2): 120-129.
| 亚组 Subgroup | 染色体 Chromosome | 染色体大小 Size of chromosome/Mb | SSR数量 Number of SSRs | SSR密度 SSR density/(SSR·Mb-1) | SSR距离 SSR distance/(kb·SSR-1) |
|---|---|---|---|---|---|
| A | 1A | 551.55 | 46 585 | 84.46 | 11.84 |
| 2A | 462.81 | 41 810 | 90.34 | 11.07 | |
| 3A | 418.65 | 34 491 | 82.39 | 12.14 | |
| 4A | 468.67 | 41 615 | 88.79 | 11.26 | |
| 5A | 489.91 | 40 918 | 83.52 | 11.97 | |
| 6A | 456.82 | 40 619 | 88.92 | 11.25 | |
| 7A | 497.83 | 41 202 | 82.76 | 12.08 | |
| C | 1C | 459.16 | 30 835 | 67.15 | 14.89 |
| 2C | 574.16 | 38 944 | 67.83 | 14.74 | |
| 3C | 616.20 | 49 132 | 79.73 | 12.54 | |
| 4C | 701.82 | 46 713 | 66.56 | 15.02 | |
| 5C | 601.56 | 41 519 | 69.02 | 14.49 | |
| 6C | 610.20 | 45 292 | 74.22 | 13.47 | |
| 7C | 530.95 | 37 060 | 69.80 | 14.33 | |
| D | 1D | 473.71 | 36 983 | 78.07 | 12.80 |
| 2D | 532.98 | 41 375 | 77.63 | 12.88 | |
| 3D | 464.06 | 33 680 | 72.58 | 13.78 | |
| 4D | 451.84 | 37 661 | 83.35 | 12.00 | |
| 5D | 500.51 | 39 941 | 79.80 | 12.53 | |
| 6D | 293.84 | 22 043 | 75.02 | 13.33 | |
| 7D | 498.92 | 39 720 | 79.61 | 12.56 | |
| 合计Total | 10 656.15 | 828 138 | - | - | |
表1 SSR数量分布特征
Table 1 SSR quantitative distribution characteristics
| 亚组 Subgroup | 染色体 Chromosome | 染色体大小 Size of chromosome/Mb | SSR数量 Number of SSRs | SSR密度 SSR density/(SSR·Mb-1) | SSR距离 SSR distance/(kb·SSR-1) |
|---|---|---|---|---|---|
| A | 1A | 551.55 | 46 585 | 84.46 | 11.84 |
| 2A | 462.81 | 41 810 | 90.34 | 11.07 | |
| 3A | 418.65 | 34 491 | 82.39 | 12.14 | |
| 4A | 468.67 | 41 615 | 88.79 | 11.26 | |
| 5A | 489.91 | 40 918 | 83.52 | 11.97 | |
| 6A | 456.82 | 40 619 | 88.92 | 11.25 | |
| 7A | 497.83 | 41 202 | 82.76 | 12.08 | |
| C | 1C | 459.16 | 30 835 | 67.15 | 14.89 |
| 2C | 574.16 | 38 944 | 67.83 | 14.74 | |
| 3C | 616.20 | 49 132 | 79.73 | 12.54 | |
| 4C | 701.82 | 46 713 | 66.56 | 15.02 | |
| 5C | 601.56 | 41 519 | 69.02 | 14.49 | |
| 6C | 610.20 | 45 292 | 74.22 | 13.47 | |
| 7C | 530.95 | 37 060 | 69.80 | 14.33 | |
| D | 1D | 473.71 | 36 983 | 78.07 | 12.80 |
| 2D | 532.98 | 41 375 | 77.63 | 12.88 | |
| 3D | 464.06 | 33 680 | 72.58 | 13.78 | |
| 4D | 451.84 | 37 661 | 83.35 | 12.00 | |
| 5D | 500.51 | 39 941 | 79.80 | 12.53 | |
| 6D | 293.84 | 22 043 | 75.02 | 13.33 | |
| 7D | 498.92 | 39 720 | 79.61 | 12.56 | |
| 合计Total | 10 656.15 | 828 138 | - | - | |
| 项目 Item | 平均值 Average | 标准差 Standard deviation | 染色体大小 Size of chromosome | SSR密度 SSR density | SSR距离 SSR distance | SSR数量 Number of SSRs |
|---|---|---|---|---|---|---|
| 染色体大小Size of chromosome | 507.44 | 85.19 | 1 | |||
| SSR密度SSR density | 78.16 | 7.38 | -0.38 | 1 | ||
| SSR距离SSR distance | 12.90 | 1.24 | 0.40 | -0.99** | 1 | |
| SSR数量Number of SSRs | 39 435.14 | 5 945.73 | 0.81** | 0.21 | -0.20 | 1 |
表2 相关性分析
Table 2 Correlation analysis
| 项目 Item | 平均值 Average | 标准差 Standard deviation | 染色体大小 Size of chromosome | SSR密度 SSR density | SSR距离 SSR distance | SSR数量 Number of SSRs |
|---|---|---|---|---|---|---|
| 染色体大小Size of chromosome | 507.44 | 85.19 | 1 | |||
| SSR密度SSR density | 78.16 | 7.38 | -0.38 | 1 | ||
| SSR距离SSR distance | 12.90 | 1.24 | 0.40 | -0.99** | 1 | |
| SSR数量Number of SSRs | 39 435.14 | 5 945.73 | 0.81** | 0.21 | -0.20 | 1 |
| 基因组Genome | 亚组Subgroup | 染色体长度Chromosome length/Mb | SSR数目Number of SSR | SSR密度SSR density/(SSR·Mb-1) |
|---|---|---|---|---|
| Sang | A | 3 277.34 | 242 707 | 73.99 |
| C | 3 867.70 | 260 585 | 67.30 | |
| D | 3 125.40 | 223 135 | 71.32 | |
| 三分三 | A | 3 346.24 | 287 240 | 85.88 |
| C | 4 094.05 | 289 495 | 70.62 | |
| D | 3 215.86 | 251 403 | 78.01 |
表3 亚基因组间SSR分布特征
Table 3 SSR distribution characteristics among subgroups
| 基因组Genome | 亚组Subgroup | 染色体长度Chromosome length/Mb | SSR数目Number of SSR | SSR密度SSR density/(SSR·Mb-1) |
|---|---|---|---|---|
| Sang | A | 3 277.34 | 242 707 | 73.99 |
| C | 3 867.70 | 260 585 | 67.30 | |
| D | 3 125.40 | 223 135 | 71.32 | |
| 三分三 | A | 3 346.24 | 287 240 | 85.88 |
| C | 4 094.05 | 289 495 | 70.62 | |
| D | 3 215.86 | 251 403 | 78.01 |
| 重复次数 Number of repeats | 重复类型Repeat type | 合计 Total | 占比 Proportion/% | |||||
|---|---|---|---|---|---|---|---|---|
| 单核苷酸 Mononucleotide | 双核苷酸 Dinucleotide | 三核苷酸 Trinucleotide | 四核苷酸 Tetranucleotide | 五核苷酸 Pentanucleotide | 六核苷酸 Hexanucleotide | |||
| 5 | - | 0 | 164 814 | 8 794 | 1 467 | 2 281 | 177 356 | 21.42 |
| 6 | - | 131 050 | 50 279 | 2 990 | 427 | 970 | 185 716 | 22.43 |
| 7 | - | 56 606 | 20 222 | 1 181 | 179 | 526 | 78 714 | 9.50 |
| 8 | - | 28 448 | 10 737 | 583 | 77 | 316 | 40 161 | 4.85 |
| 9 | - | 14 827 | 6 875 | 361 | 49 | 207 | 22 319 | 2.70 |
| 10 | - | 7 751 | 4 958 | 223 | 26 | 165 | 13 123 | 1.58 |
| >10 | 247 167 | 40 281 | 21 936 | 816 | 96 | 453 | 310 749 | 37.52 |
| 合计Total | 247 167 | 278 963 | 279 821 | 14 948 | 2 321 | 4 918 | 828 138 | - |
| 占比Proportion /% | 29.85 | 33.69 | 33.79 | 1.81 | 0.28 | 0.59 | - | 100 |
表4 燕麦全基因组SSR重复类型、重复次数和占比
Table 4 SSR repeat type, repeat number and proportion of whole genome of oat
| 重复次数 Number of repeats | 重复类型Repeat type | 合计 Total | 占比 Proportion/% | |||||
|---|---|---|---|---|---|---|---|---|
| 单核苷酸 Mononucleotide | 双核苷酸 Dinucleotide | 三核苷酸 Trinucleotide | 四核苷酸 Tetranucleotide | 五核苷酸 Pentanucleotide | 六核苷酸 Hexanucleotide | |||
| 5 | - | 0 | 164 814 | 8 794 | 1 467 | 2 281 | 177 356 | 21.42 |
| 6 | - | 131 050 | 50 279 | 2 990 | 427 | 970 | 185 716 | 22.43 |
| 7 | - | 56 606 | 20 222 | 1 181 | 179 | 526 | 78 714 | 9.50 |
| 8 | - | 28 448 | 10 737 | 583 | 77 | 316 | 40 161 | 4.85 |
| 9 | - | 14 827 | 6 875 | 361 | 49 | 207 | 22 319 | 2.70 |
| 10 | - | 7 751 | 4 958 | 223 | 26 | 165 | 13 123 | 1.58 |
| >10 | 247 167 | 40 281 | 21 936 | 816 | 96 | 453 | 310 749 | 37.52 |
| 合计Total | 247 167 | 278 963 | 279 821 | 14 948 | 2 321 | 4 918 | 828 138 | - |
| 占比Proportion /% | 29.85 | 33.69 | 33.79 | 1.81 | 0.28 | 0.59 | - | 100 |
| 重复类型 Repeat type | 重复数目 Number of repeats | 优势重复基元 Dominant repeat primitives | SSR数目 Number of SSR | 占本重复类型百分比 Percentage of predominant motifs/% |
|---|---|---|---|---|
| 单核苷酸 Mononucleotide | 4 | A/T | 145 340 | 58.80 |
| C/G | 101 827 | 41.20 | ||
| 双核苷酸 Dinucleotide | 12 | AG/CT | 137 111 | 49.15 |
| AT/AT | 69 342 | 24.86 | ||
| AC/GT | 67 856 | 24.32 | ||
| CG/CG | 4 654 | 1.67 | ||
| 三核苷酸 Trinucleotide | 60 | AAC/GTT | 68 530 | 24.49 |
| ATC/ATG | 57 361 | 20.50 | ||
| AAG/CTT | 45 805 | 16.37 | ||
| AGG/CCT | 28 650 | 10.24 | ||
| 其他 Others | 79 475 | 28.40 | ||
| 四核苷酸 Tetranucleotide | 238 | TTCA/TGAA | 3 968 | 26.55 |
| AAAT/ATTT | 2 286 | 15.29 | ||
| ATAG/CTAT | 1 753 | 11.73 | ||
| AAAG/CTTT | 1 003 | 6.71 | ||
| 其他 Others | 5 938 | 39.72 | ||
| 五核苷酸 Pentanucleotide | 390 | TAAGA/TCTTA | 910 | 39.21 |
| CCAAA/TTTGG | 186 | 8.01 | ||
| CGAGG/CCTCG | 176 | 7.58 | ||
| CGCCA/TGGC | 126 | 5.43 | ||
| 其他 Others | 923 | 39.77 | ||
| 六核苷酸 Hexanucleotide | 844 | CCTGGG/CCCAGG | 2 053 | 41.91 |
| AGGAGA/TCTCCT | 147 | 3.00 | ||
| TTGTTA/TAACAA | 142 | 2.90 | ||
| TTTAAA/TTTAAA | 130 | 2.65 | ||
| 其他 Others | 2 427 | 49.54 |
表5 燕麦全基因组各SSR重复类型中优势基元类型及分布
Table 5 Types and distribution of dominant motifs in each SSR repeat type of oat genome
| 重复类型 Repeat type | 重复数目 Number of repeats | 优势重复基元 Dominant repeat primitives | SSR数目 Number of SSR | 占本重复类型百分比 Percentage of predominant motifs/% |
|---|---|---|---|---|
| 单核苷酸 Mononucleotide | 4 | A/T | 145 340 | 58.80 |
| C/G | 101 827 | 41.20 | ||
| 双核苷酸 Dinucleotide | 12 | AG/CT | 137 111 | 49.15 |
| AT/AT | 69 342 | 24.86 | ||
| AC/GT | 67 856 | 24.32 | ||
| CG/CG | 4 654 | 1.67 | ||
| 三核苷酸 Trinucleotide | 60 | AAC/GTT | 68 530 | 24.49 |
| ATC/ATG | 57 361 | 20.50 | ||
| AAG/CTT | 45 805 | 16.37 | ||
| AGG/CCT | 28 650 | 10.24 | ||
| 其他 Others | 79 475 | 28.40 | ||
| 四核苷酸 Tetranucleotide | 238 | TTCA/TGAA | 3 968 | 26.55 |
| AAAT/ATTT | 2 286 | 15.29 | ||
| ATAG/CTAT | 1 753 | 11.73 | ||
| AAAG/CTTT | 1 003 | 6.71 | ||
| 其他 Others | 5 938 | 39.72 | ||
| 五核苷酸 Pentanucleotide | 390 | TAAGA/TCTTA | 910 | 39.21 |
| CCAAA/TTTGG | 186 | 8.01 | ||
| CGAGG/CCTCG | 176 | 7.58 | ||
| CGCCA/TGGC | 126 | 5.43 | ||
| 其他 Others | 923 | 39.77 | ||
| 六核苷酸 Hexanucleotide | 844 | CCTGGG/CCCAGG | 2 053 | 41.91 |
| AGGAGA/TCTCCT | 147 | 3.00 | ||
| TTGTTA/TAACAA | 142 | 2.90 | ||
| TTTAAA/TTTAAA | 130 | 2.65 | ||
| 其他 Others | 2 427 | 49.54 |
| 引物 Primer | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') | 退火温度Annealing temperature/℃ | 在4D染色体起始位置Starting position/bp | 扩增长度Amplification length/bp | 基元类型Primitive type | 重复次数Number of repeats |
|---|---|---|---|---|---|---|---|
| A-1 | ATGGTAAACGGGTCATCGAGT | AAGTATAGGCCGACGATCCAG | 54 | 402 056 122 | 195 | TC | 27 |
| A-2 | TCCTTAGGCCGTGAAGAAGTT | TGCTCTATGTACTGATCCGTGA | 54 | 403 098 125 | 184 | GA | 12 |
| A-3 | CCCATAGTAGACCCTTGCCAT | ACGAGAGACTGAAGTGAAGCT | 58 | 404 060 755 | 296 | TTC | 9 |
| A-4 | TATACAATGGACCTCGGCAGA | ACTGCGTACACTGCTAGCTAT | 58 | 413 048 093 | 227 | TA | 30 |
| A-5 | ATTCCGGACTCTGCTTACGAT | TATCCTCAGGTATGCGTGGTT | 58 | 414 065 870 | 232 | TACA | 20 |
| A-6 | TGCATAGTGTTACCAGCGAAC | CGGTTTGAAAGATAGGGGTGG | 58 | 415 521 048 | 291 | TA | 26 |
| A-7 | CTGCTCACCACCACAATTCAA | TGAGCTATTCTACCACGTCCC | 58 | 416 396 796 | 292 | TA | 22 |
| A-8 | TCCTTAGGCCGTGAAGAAGTT | TGCTCTATGTACTGATCCGTGA | 58 | 417 151 310 | 243 | GA | 52 |
| A-9 | ATGGTAAACGGGTCATCGAGT | AAGTATAGGCCGACGATCCAG | 59 | 418 130 363 | 204 | TC | 39 |
| A-10 | CACTGCCTGGTAAACTTCGAT | TGCTCATGTCTATTAACGTCCA | 57 | 420 675 833 | 289 | ATT | 11 |
| A-11 | CTCAGGTGCAGAGGAGAACTT | TGTTGTGTCTTGAGCCTGTTG | 58 | 421 639 959 | 275 | GTT | 7 |
| A-12 | AGACAGAGCGAGTACTTGGAA | ATGGATGTGTGCACGTTTCAT | 58 | 422 813 420 | 231 | GATG | 8 |
| A-13 | AGTCTTTGAGTCCGTGTCCAT | ATGTGGAGGAGCAATGAAAGC | 58 | 423 221 780 | 276 | TCC | 12 |
| A-14 | AGGAGATCAATCGCGGTACAT | TGTCACTATCCCTGTCACTGG | 58 | 424 040 249 | 284 | AAG | 13 |
| A-15 | TCTAGCTTGCTGGTCCTTGTT | ACTAATTGACAACACCAGCCC | 58 | 424 831 385 | 271 | GAT | 9 |
| A-16 | TGAAATCTGCCGGGGAAATTC | AGTAGTCAACACCGGGAAAGT | 58 | 424 944 085 | 200 | ATC | 5 |
| A-17 | GACTTCTAGGTCGCTGGTTC | ACAAGCGAAGGAAGAATCAC | 56 | 424 853 458 | 253 | AG | 19 |
| A-18 | GTCGTTGTAATCCCGATTGT | CTTCCTCCCTGAGTTCGATA | 57 | 424 714 810 | 391 | TA | 38 |
| A-19 | TTCAAACTATGGGTGCACTT | GATGTAAGACTGGCCTCCTG | 57 | 424 623 031 | 362 | TA | 26 |
| A-20 | AGCGTGTTATGTGGTGTTTG | CCGTCGCGTAGAATAAAAGT | 57 | 424 595 914 | 349 | AT | 27 |
| A-21 | CGATGTTGTGAGGGACAATA | ATCGACTGGACAAATGGAGT | 57 | 423 715 272 | 364 | TA | 30 |
| A-22 | CTCTGACAGTCCAACTGCAA | AGTCGTCGTATGTTGTGTGG | 57 | 423 425 714 | 333 | CAA | 10 |
| A-23 | TTGGTGGTCTCTTGCTGTTA | TGGGCCAAAGAGAATTTTAG | 57 | 423 121 188 | 307 | TA | 49 |
| A-24 | TGATCCACCTCCAGGACTAT | CCATCTCTAGGCCTTCACTG | 57 | 422 863 900 | 394 | GA | 26 |
| A-25 | TATCATCGAGTTCCGACTCA | TAGTGTCCTGCATGGTTGTC | 57 | 422 092 205 | 344 | TTC | 10 |
| A-26 | AGTGGTGGGAGTGAGGTGT | CACCTCCAAGCCCTAAACTA | 57 | 422 030 145 | 260 | GA | 81 |
| A-27 | GGAAAACACCGAGAGAGAAA | TGTCCTCAAGAACAAAACGA | 57 | 421 993 919 | 378 | ATC | 8 |
| A-28 | ACTAAAGGTTGCTCCGTTTG | TGAGCTTGCACATCATTTCT | 57 | 421 827 958 | 331 | AGA | 6 |
| A-29 | CTCCCTCCGTTTTGAAAATA | CCATCAACGGAACATGTAAA | 57 | 420 311 577 | 366 | AT | 30 |
| A-30 | TGTGGTGAATATGCCTCTTG | AAAGGAAAAGGGAGAAAGGA | 57 | 420 091 310 | 369 | AG | 30 |
| A-31 | GGAGTCGTTCTTGATGTTGA | ATAATCCAATCCAACCCAAA | 57 | 419 525 753 | 365 | CGA | 9 |
| A-32 | AAACCTGACTGGACCTTCTG | GATGGCAATCCAACAGTTTT | 57 | 418 055 976 | 265 | TA | 27 |
| A-33 | ACTTTGGTTGCTCTCTTTGC | GAAACCTCCGTATTGTCGAT | 57 | 417 823 067 | 398 | AG | 43 |
| A-34 | ACTTTGGTTGCTCTCTTTGC | GAAACCTCCGTATTGTCGAT | 57 | 417 740 723 | 393 | AT | 36 |
| A-35 | TGGATCACTCATACGGTCAG | GAGGGGTCATTGTCTACAGG | 57 | 416 846 114 | 391 | GA | 46 |
| A-36 | GGTAAACGCAAAGCTGAAAT | AAAGGGCATGGTATCCATTA | 57 | 416 404 269 | 324 | GA | 6 |
| A-37 | TCGAAAGGGAATGTTCTAGC | TTGCTCCTACTGCCACATTA | 57 | 415 943 775 | 312 | CT | 20 |
| A-38 | TTTGGGAGGACCTCTAGTTG | TACCCTTGAAGACCGAAAAG | 57 | 415 647 138 | 370 | AAC | 5 |
| A-39 | ATGCAACAAATGATGAATGC | TAGTGCGAAAACCATCTTCA | 57 | 414 940 663 | 394 | CT | 16 |
| A-40 | CTAACTGAGCGTCGCATAAC | CTCAAGGGATGGGGATAAG | 57 | 414 641 081 | 351 | AT | 30 |
| A-41 | TGTTCATGTATCCCGCATAG | TTCCAGACGCCTAGGTTTAG | 57 | 414 586 340 | 359 | AT | 31 |
| A-42 | TGAAAGATAAGGGGTGGATG | AAGTGCACCCATAGTTTGAA | 57 | 414 263 854 | 378 | TA | 13 |
| A-43 | GCCATTATTTGTGCATGAAG | ATGTGGGGTGCAAAATAACT | 57 | 413 518 395 | 355 | TA | 34 |
| A-44 | AGTGCATCGAACTCTCTTCC | GCTGCGTACGTACATCTCAA | 57 | 413 420 890 | 337 | TC | 46 |
| A-45 | TGATGAATCGAGAAGACACG | ATTCGCAACATGAAAAACAA | 57 | 413 194 121 | 377 | ATG | 13 |
| A-46 | CAAGATGCGGATGATGATTA | GAACCTTCGATACCACAACC | 57 | 412 897 229 | 365 | TC | 6 |
| A-47 | ATGACTTTCCGCAATCTCTC | AAGGTTTGACACGATGACCT | 57 | 411 869 015 | 316 | TTC | 24 |
| A-48 | CCCAAATAGACCAAGGAATG | ATACTGAGCAGGCAAAGGAC | 57 | 411 852 406 | 370 | AT | 40 |
| A-49 | AAAAGCCCTGACTAAGAGCA | TTCCTTTGGTCTCAAAATGG | 57 | 409 342 863 | 396 | TA | 40 |
| A-50 | GGGAAAGATCAATGGTTCAG | TTCCAAAAGCTGACACGTTA | 57 | 408 678 582 | 259 | TA | 33 |
| A-51 | GGTGGTTAGATGGGATCAGA | ACTCTGGGTGTTGAATAGGG | 57 | 406 013 380 | 347 | TC | 31 |
| A-52 | TAGAATTCCTCCCACATGGT | CGATCCTTGCAGTTAGGAGT | 57 | 405 122 840 | 239 | GA | 12 |
表6 开发的燕麦多态性SSR引物信息
Table 6 Information of SSR primers developed for oats polymorphism
| 引物 Primer | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') | 退火温度Annealing temperature/℃ | 在4D染色体起始位置Starting position/bp | 扩增长度Amplification length/bp | 基元类型Primitive type | 重复次数Number of repeats |
|---|---|---|---|---|---|---|---|
| A-1 | ATGGTAAACGGGTCATCGAGT | AAGTATAGGCCGACGATCCAG | 54 | 402 056 122 | 195 | TC | 27 |
| A-2 | TCCTTAGGCCGTGAAGAAGTT | TGCTCTATGTACTGATCCGTGA | 54 | 403 098 125 | 184 | GA | 12 |
| A-3 | CCCATAGTAGACCCTTGCCAT | ACGAGAGACTGAAGTGAAGCT | 58 | 404 060 755 | 296 | TTC | 9 |
| A-4 | TATACAATGGACCTCGGCAGA | ACTGCGTACACTGCTAGCTAT | 58 | 413 048 093 | 227 | TA | 30 |
| A-5 | ATTCCGGACTCTGCTTACGAT | TATCCTCAGGTATGCGTGGTT | 58 | 414 065 870 | 232 | TACA | 20 |
| A-6 | TGCATAGTGTTACCAGCGAAC | CGGTTTGAAAGATAGGGGTGG | 58 | 415 521 048 | 291 | TA | 26 |
| A-7 | CTGCTCACCACCACAATTCAA | TGAGCTATTCTACCACGTCCC | 58 | 416 396 796 | 292 | TA | 22 |
| A-8 | TCCTTAGGCCGTGAAGAAGTT | TGCTCTATGTACTGATCCGTGA | 58 | 417 151 310 | 243 | GA | 52 |
| A-9 | ATGGTAAACGGGTCATCGAGT | AAGTATAGGCCGACGATCCAG | 59 | 418 130 363 | 204 | TC | 39 |
| A-10 | CACTGCCTGGTAAACTTCGAT | TGCTCATGTCTATTAACGTCCA | 57 | 420 675 833 | 289 | ATT | 11 |
| A-11 | CTCAGGTGCAGAGGAGAACTT | TGTTGTGTCTTGAGCCTGTTG | 58 | 421 639 959 | 275 | GTT | 7 |
| A-12 | AGACAGAGCGAGTACTTGGAA | ATGGATGTGTGCACGTTTCAT | 58 | 422 813 420 | 231 | GATG | 8 |
| A-13 | AGTCTTTGAGTCCGTGTCCAT | ATGTGGAGGAGCAATGAAAGC | 58 | 423 221 780 | 276 | TCC | 12 |
| A-14 | AGGAGATCAATCGCGGTACAT | TGTCACTATCCCTGTCACTGG | 58 | 424 040 249 | 284 | AAG | 13 |
| A-15 | TCTAGCTTGCTGGTCCTTGTT | ACTAATTGACAACACCAGCCC | 58 | 424 831 385 | 271 | GAT | 9 |
| A-16 | TGAAATCTGCCGGGGAAATTC | AGTAGTCAACACCGGGAAAGT | 58 | 424 944 085 | 200 | ATC | 5 |
| A-17 | GACTTCTAGGTCGCTGGTTC | ACAAGCGAAGGAAGAATCAC | 56 | 424 853 458 | 253 | AG | 19 |
| A-18 | GTCGTTGTAATCCCGATTGT | CTTCCTCCCTGAGTTCGATA | 57 | 424 714 810 | 391 | TA | 38 |
| A-19 | TTCAAACTATGGGTGCACTT | GATGTAAGACTGGCCTCCTG | 57 | 424 623 031 | 362 | TA | 26 |
| A-20 | AGCGTGTTATGTGGTGTTTG | CCGTCGCGTAGAATAAAAGT | 57 | 424 595 914 | 349 | AT | 27 |
| A-21 | CGATGTTGTGAGGGACAATA | ATCGACTGGACAAATGGAGT | 57 | 423 715 272 | 364 | TA | 30 |
| A-22 | CTCTGACAGTCCAACTGCAA | AGTCGTCGTATGTTGTGTGG | 57 | 423 425 714 | 333 | CAA | 10 |
| A-23 | TTGGTGGTCTCTTGCTGTTA | TGGGCCAAAGAGAATTTTAG | 57 | 423 121 188 | 307 | TA | 49 |
| A-24 | TGATCCACCTCCAGGACTAT | CCATCTCTAGGCCTTCACTG | 57 | 422 863 900 | 394 | GA | 26 |
| A-25 | TATCATCGAGTTCCGACTCA | TAGTGTCCTGCATGGTTGTC | 57 | 422 092 205 | 344 | TTC | 10 |
| A-26 | AGTGGTGGGAGTGAGGTGT | CACCTCCAAGCCCTAAACTA | 57 | 422 030 145 | 260 | GA | 81 |
| A-27 | GGAAAACACCGAGAGAGAAA | TGTCCTCAAGAACAAAACGA | 57 | 421 993 919 | 378 | ATC | 8 |
| A-28 | ACTAAAGGTTGCTCCGTTTG | TGAGCTTGCACATCATTTCT | 57 | 421 827 958 | 331 | AGA | 6 |
| A-29 | CTCCCTCCGTTTTGAAAATA | CCATCAACGGAACATGTAAA | 57 | 420 311 577 | 366 | AT | 30 |
| A-30 | TGTGGTGAATATGCCTCTTG | AAAGGAAAAGGGAGAAAGGA | 57 | 420 091 310 | 369 | AG | 30 |
| A-31 | GGAGTCGTTCTTGATGTTGA | ATAATCCAATCCAACCCAAA | 57 | 419 525 753 | 365 | CGA | 9 |
| A-32 | AAACCTGACTGGACCTTCTG | GATGGCAATCCAACAGTTTT | 57 | 418 055 976 | 265 | TA | 27 |
| A-33 | ACTTTGGTTGCTCTCTTTGC | GAAACCTCCGTATTGTCGAT | 57 | 417 823 067 | 398 | AG | 43 |
| A-34 | ACTTTGGTTGCTCTCTTTGC | GAAACCTCCGTATTGTCGAT | 57 | 417 740 723 | 393 | AT | 36 |
| A-35 | TGGATCACTCATACGGTCAG | GAGGGGTCATTGTCTACAGG | 57 | 416 846 114 | 391 | GA | 46 |
| A-36 | GGTAAACGCAAAGCTGAAAT | AAAGGGCATGGTATCCATTA | 57 | 416 404 269 | 324 | GA | 6 |
| A-37 | TCGAAAGGGAATGTTCTAGC | TTGCTCCTACTGCCACATTA | 57 | 415 943 775 | 312 | CT | 20 |
| A-38 | TTTGGGAGGACCTCTAGTTG | TACCCTTGAAGACCGAAAAG | 57 | 415 647 138 | 370 | AAC | 5 |
| A-39 | ATGCAACAAATGATGAATGC | TAGTGCGAAAACCATCTTCA | 57 | 414 940 663 | 394 | CT | 16 |
| A-40 | CTAACTGAGCGTCGCATAAC | CTCAAGGGATGGGGATAAG | 57 | 414 641 081 | 351 | AT | 30 |
| A-41 | TGTTCATGTATCCCGCATAG | TTCCAGACGCCTAGGTTTAG | 57 | 414 586 340 | 359 | AT | 31 |
| A-42 | TGAAAGATAAGGGGTGGATG | AAGTGCACCCATAGTTTGAA | 57 | 414 263 854 | 378 | TA | 13 |
| A-43 | GCCATTATTTGTGCATGAAG | ATGTGGGGTGCAAAATAACT | 57 | 413 518 395 | 355 | TA | 34 |
| A-44 | AGTGCATCGAACTCTCTTCC | GCTGCGTACGTACATCTCAA | 57 | 413 420 890 | 337 | TC | 46 |
| A-45 | TGATGAATCGAGAAGACACG | ATTCGCAACATGAAAAACAA | 57 | 413 194 121 | 377 | ATG | 13 |
| A-46 | CAAGATGCGGATGATGATTA | GAACCTTCGATACCACAACC | 57 | 412 897 229 | 365 | TC | 6 |
| A-47 | ATGACTTTCCGCAATCTCTC | AAGGTTTGACACGATGACCT | 57 | 411 869 015 | 316 | TTC | 24 |
| A-48 | CCCAAATAGACCAAGGAATG | ATACTGAGCAGGCAAAGGAC | 57 | 411 852 406 | 370 | AT | 40 |
| A-49 | AAAAGCCCTGACTAAGAGCA | TTCCTTTGGTCTCAAAATGG | 57 | 409 342 863 | 396 | TA | 40 |
| A-50 | GGGAAAGATCAATGGTTCAG | TTCCAAAAGCTGACACGTTA | 57 | 408 678 582 | 259 | TA | 33 |
| A-51 | GGTGGTTAGATGGGATCAGA | ACTCTGGGTGTTGAATAGGG | 57 | 406 013 380 | 347 | TC | 31 |
| A-52 | TAGAATTCCTCCCACATGGT | CGATCCTTGCAGTTAGGAGT | 57 | 405 122 840 | 239 | GA | 12 |
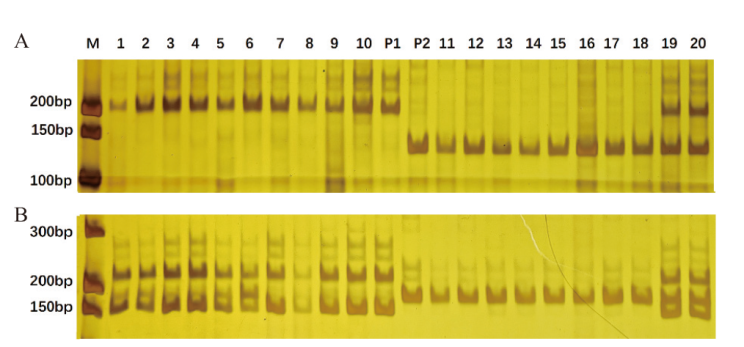
图2 引物A-1(A)和A-2(B)在试验材料中扩增结果 M:DNA marker;P1:Banner;P2:白燕二号;1-20:20株F2单株
Fig. 2 Amplification results of primers A-1(A)and A-2(B)in experimental materials M: DNA marker; P1: Banner; P2: Baiyan 2; 1-20: F2 20 individual plants
| [1] | 吴斌, 郑殿升, 严威凯, 等. 燕麦分子育种研究进展[J]. 植物遗传资源学报, 2019, 20(3): 485-495. |
| Wu B, Zheng DS, Yan KW, et al. Advances in molecular breeding of oats[J]. Journal of Plant Genetic Resources, 2019, 20(3): 485-495. | |
| [2] | Rasane P, Jha A, Sabikhi L, et al. Nutritional advantages of oats and opportunities for its processing as value added foods-areview[J]. J Food Sci Technol, 2015, 52(2): 662-675. |
| [3] |
潘莹, 程时锋. 燕麦基因组学研究进展[J]. 植物遗传资源学报, 2021, 22(2): 304-308.
doi: 10.13430/j.cnki.jpgr.20200910001 |
| Pan Y, Cheng SF. Research progress on oat genomics study[J]. J Plant Genet Res, 2021, 22(2): 304-308. | |
| [4] |
Kamal N, Tsardakas Renhuldt N, Bentzer J, et al. The mosaic oat genome gives insights into a uniquely healthy cereal crop[J]. Nature, 2022, 606(7912): 113-119.
doi: 10.1038/s41586-022-04732-y |
| [5] |
Peng YY, Yan HH, Guo LC, et al. Reference genome assemblies reveal the origin and evolution of allohexaploid oat[J]. Nat Genet, 2022, 54(8): 1248-1258.
doi: 10.1038/s41588-022-01127-7 |
| [6] | Wu JZ, Zhao Q, Wu GW, et al. Development of novel SSR markers for flax(Linum usitatissimum L.)using reduced-representation genome sequencing[J]. Front Plant Sci, 2017, 7: 2018. |
| [7] |
Guo R, Landis JB, Moore MJ, et al. Development and application of transcriptome-derived microsatellites in Actinidia eriantha(Actinidiaceae)[J]. Front Plant Sci, 2017, 8: 1383.
doi: 10.3389/fpls.2017.01383 URL |
| [8] |
Malausa T, Gilles A, Meglecz E, et al. High-throughput microsatellite isolation through 454 GS-FLX titanium pyrosequencing of enriched DNA libraries[J]. Mol Ecol Resour, 2011, 11(4): 638-644.
doi: 10.1111/j.1755-0998.2011.02992.x pmid: 21676194 |
| [9] |
Sowa S, Paczos-Grzęda E. Identification of molecular markers for the PC39 gene conferring resistance to crown rust in oat[J]. Theor Appl Genet, 2020, 133(4): 1081-1094.
doi: 10.1007/s00122-020-03533-z pmid: 31927607 |
| [10] |
Zhao J, Kebede AZ, Menzies JG, et al. Chromosomal location of the crown rust resistance gene Pc98 in cultivated oat(Avena sativa L.)[J]. Theor Appl Genet, 2020, 133(4): 1109-1122.
doi: 10.1007/s00122-020-03535-x pmid: 31938813 |
| [11] |
Yan H, Yu K, Xu Y, et al. Position validation of the dwarfing gene dw6 in oat(Avena sativa L.)and its correlated effects on agronomic traits[J]. Front Plant Sci, 2021, 12: 668847.
doi: 10.3389/fpls.2021.668847 URL |
| [12] |
吴斌, 张茜, 宋高原, 等. 裸燕麦SSR标记连锁群图谱的构建及β-葡聚糖含量QTL的定位[J]. 中国农业科学, 2014, 47(6): 1208-1215.
doi: 10.3864/j.issn.0578-1752.2014.06.017 |
| Wu B, Zhang Q, Song GY, et al. Construction of SSR genetic linkage map and analysis of QTLs related to β-glucan Content of naked oat(Avena nuda L.)[J]. Sci Agric Sin, 2014, 47(6): 1208-1215. | |
| [13] |
Isabel LP, Park JR, Lee GS, et al. Development of EST-SSR markers and analysis of genetic relationship it's resources in hexaploid oats[J]. J Crop Sci Biotechnol, 2019, 22(3): 243-251.
doi: 10.1007/s12892-019-0158-0 |
| [14] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [15] |
Ubert IP, Zimmer CM, Pellizzaro K, et al. Genetics and molecular mapping of the naked grains in hexaploid oat[J]. Euphytica, 2017, 213(2): 41.
doi: 10.1007/s10681-017-1836-1 |
| [16] |
Zhou Y, Zhao XB, Li YW, et al. Triticum population sequencing provides insights into wheat adaptation[J]. Nat Genet, 2020, 52(12): 1412-1422.
doi: 10.1038/s41588-020-00722-w pmid: 33106631 |
| [17] | 原志敏. 玉米全基因组SSRs分子标记开发与特征分析[D]. 雅安: 四川农业大学, 2013. |
| Yuan ZM. Development and characterization of SSRs molecular markers in maize genome[D]. Ya'an: Sichuan Agricultural University, 2013. | |
| [18] | 马名川, 刘龙龙, 刘璋, 等. 苦荞全基因组SSR位点特征分析与分子标记开发[J]. 作物杂志, 2021(1): 38-46. |
| Ma MC, Liu LL, Liu Z, et al. Analysis of SSR loci in whole genome and development of molecular markers in tartary buckwheat[J]. Crops, 2021(1): 38-46. | |
| [19] | 张晗, 王雪梅, 王东建, 等. 谷子基因组SSR信息分析和标记开发[J]. 分子植物育种, 2013, 11(1): 30-36. |
| Zhang H, Wang XM, Wang DJ, et al. Survey of SSRs in foxtail millet genome and development of SSR markers[J]. Mol Plant Breed, 2013, 11(1): 30-36. | |
| [20] | 仇静静. 野生花生全基因组SSR标记的开发与应用[D]. 济南: 山东师范大学, 2018. |
| QIU JJ. Development and application of SSR markers in wild peanut genome[D]. Jinan: Shandong Normal University, 2018. | |
| [21] |
Cavagnaro PF, Senalik DA, Yang LM, et al. Genome-wide characterization of simple sequence repeats in cucumber(Cucumis sativus L.)[J]. BMC Genomics, 2010, 11: 569.
doi: 10.1186/1471-2164-11-569 pmid: 20950470 |
| [22] |
关玲, 章镇, 王新卫, 等. 苹果基因组SSR位点分析与应用[J]. 中国农业科学, 2011, 44(21): 4415-4428.
doi: 10.3864/j.issn.0578-1752.2011.21.010 |
|
Guan L, Zhang Z, Wang XW, et al. Evaluation and application of the SSR loci in apple genome[J]. Sci Agric Sin, 2011, 44(21): 4415-4428.
doi: 10.3864/j.issn.0578-1752.2011.21.010 |
|
| [23] |
李乔乔, 王宇晴, 刘蕊, 等. 甜菜全基因组SSR引物的筛选与评价[J]. 中国农学通报, 2022, 38(12): 95-99.
doi: 10.11924/j.issn.1000-6850.casb2021-0726 |
|
Li QQ, Wang YQ, Liu R, et al. The whole genome SSR primers of sugar beet: screening and evaluation[J]. Chin Agric Sci Bull, 2022, 38(12): 95-99.
doi: 10.11924/j.issn.1000-6850.casb2021-0726 |
|
| [24] |
Han B, Wang CB, Tang ZH, et al. Genome-wide analysis of microsatellite markers based on sequenced database in chinese spring wheat(Triticum aestivum L.)[J]. PLoS One, 2015, 10(11): e0141540.
doi: 10.1371/journal.pone.0141540 URL |
| [25] |
Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes[J]. Nat Genet, 2002, 30(2): 194-200.
doi: 10.1038/ng822 pmid: 11799393 |
| [26] |
张霞, 于卓, 金兴红, 等. 马铃薯SSR引物的开发、特征分析及在彩色马铃薯材料中的扩增研究[J]. 作物学报, 2022, 48(4): 920-929.
doi: 10.3724/SP.J.1006.2022.14065 |
|
Zhang X, Yu Z, Jin XH, et al. Development and characterization analysis of potato SSR primers and the amplification research in colored potato materials[J]. Acta Agron Sin, 2022, 48(4): 920-929.
doi: 10.3724/SP.J.1006.2022.14065 |
|
| [27] | 杜磊, 蒙秋伊, 尚昆, 等. 薏苡基因组SSR标记开发与应用[J]. 分子植物育种, 2022, 20(3): 887-894. |
| Du L, Meng QY, Shang K, et al. Development and application of genomic SSR markers in coix lachryma-jobi genome[J]. Mol Plant Breed, 2022, 20(3): 887-894. | |
| [28] |
姚嘉瑜, 张立武, 赵捷, 等. 黄麻全基因组SSR鉴定与特征分析[J]. 作物学报, 2019, 45(1): 10-17.
doi: 10.3724/SP.J.1006.2019.84072 |
|
Yao JY, Zhang LW, Zhao J, et al. Evaluation and characteristic analysis of SSRs from the whole genome of jute(Corchorus capsularis)[J]. Acta Agron Sin, 2019, 45(1): 10-17.
doi: 10.3724/SP.J.1006.2019.84072 URL |
|
| [29] |
童治军, 焦芳婵, 肖炳光. 普通烟草及其祖先种基因组SSR位点分析[J]. 中国农业科学, 2015, 48(11): 2108-2117.
doi: 10.3864/j.issn.0578-1752.2015.11.003 |
| Tong ZJ, Jiao FC, Xiao BG. Analysis of SSR loci in nicotina tabacum genome and its two ancestral species genome[J]. Sci Agric Sin, 2015, 48(11): 2108-2117. | |
| [30] |
叶卫军, 陈圣男, 杨勇, 等. 绿豆SSR标记的开发及遗传多样性分析[J]. 作物学报, 2019, 45(8): 1176-1188.
doi: 10.3724/SP.J.1006.2019.84155 |
| Ye WJ, Chen SN, Yang Y, et al. Development of SSR markers and genetic diversity analysis in mung bean[J]. Acta Agron Sin, 2019, 45(8): 1176-1188. | |
| [31] |
Oliveira EJ, Pádua JG, Zucchi MI, et al. Origin, evolution and genome distribution of microsatellites[J]. Genet Mol Biol, 2006, 29(2): 294-307.
doi: 10.1590/S1415-47572006000200018 URL |
| [32] |
Luo MC, Gu YQ, Puiu D, et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii[J]. Nature, 2017, 551(7681): 498-502.
doi: 10.1038/nature24486 URL |
| [1] | 张岳一, 兰社益, 裴海闰, 封棣. 多菌种联用发酵燕麦麸皮工艺优化及发用功效评价[J]. 生物技术通报, 2023, 39(9): 58-70. |
| [2] | 李琦, 杨晓蕾, 李晓林, 申友磊, 李建宏, 姚拓. 高寒草地燕麦根际解植酸磷促生菌鉴定及其优势菌假单胞菌属菌株功能特性[J]. 生物技术通报, 2023, 39(3): 243-253. |
| [3] | 余世洲, 曹领改, 王世泽, 刘勇, 边文杰, 任学良. 烟草种质基因分型核心SNP标记的开发[J]. 生物技术通报, 2023, 39(3): 89-100. |
| [4] | 安苗, 王彤彤, 付逸婷, 夏俊俊, 彭锁堂, 段永红. 52个马铃薯遗传多样性分析及SSR分子身份证构建[J]. 生物技术通报, 2023, 39(12): 136-147. |
| [5] | 符勇耀, 易德燕, 杨先茂, 蔡莉, 梁渝华, 雷美艳, 杨利平. 卷丹新种质JD-h-15的形态特征与遗传变异分析[J]. 生物技术通报, 2022, 38(11): 140-150. |
| [6] | 张廷焕, 张利娟, 陈四清, 郭宗义. 猪miR-378种子序列的多态性对其功能以及胴体性状的影响[J]. 生物技术通报, 2021, 37(6): 154-162. |
| [7] | 管俊娇, 杨晓洪, 张鹏, 黄清梅, 张建华. 基于荧光检测技术的青稞品种鉴定方法的建立[J]. 生物技术通报, 2021, 37(11): 293-302. |
| [8] | 郭丽丽, 李昱莹, 郭大龙, 侯小改. 重要花卉植物高密度遗传连锁图谱构建研究进展[J]. 生物技术通报, 2021, 37(1): 246-254. |
| [9] | 张德荣, 马晓霞, 李羽翡, 赵永清, 霍生东, 马忠仁, 柏家林. 脂联素及其受体在哺乳动物中的研究进展与展望[J]. 生物技术通报, 2020, 36(6): 236-244. |
| [10] | 余钧剑, 迟美丽, 贾永义, 刘士力, 竺俊全, 顾志敏. 四引物扩增受阻突变体系PCR技术及其在动植物遗传育种研究中的应用[J]. 生物技术通报, 2020, 36(5): 32-38. |
| [11] | 李晓凯, 范一星, 乔贤, 张磊, 王凤红, 王志英, 王瑞军, 张燕军, 刘志红, 王志新, 何利兵, 李金泉, 苏蕊, 张家新. 山羊基因组与遗传变异图谱研究进展[J]. 生物技术通报, 2020, 36(4): 175-184. |
| [12] | 赵阳阳, 郭雨潇, 张凌云. 文冠果果实转录组测序及分析[J]. 生物技术通报, 2019, 35(6): 24-31. |
| [13] | 李标, 张瑞莹, 王小琪, 张存芳, 段子渊. 滩羊微卫星标记多态性及与体尺性状关联分析[J]. 生物技术通报, 2019, 35(6): 131-137. |
| [14] | 黄龙, 吴本丽, 何吉祥, 陈静, 宋光同, 汪翔, 张烨, 武松. 中华鳖MyoD1基因SNP鉴定及其与生长性状的关联分析[J]. 生物技术通报, 2019, 35(4): 76-81. |
| [15] | 刘培培, 罗光明, 柴华文, 张俊逸, 朱梓豪, 王得运. SSR标记技术在中药鉴定中的应用[J]. 生物技术通报, 2019, 35(2): 198-203. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||