








生物技术通报 ›› 2024, Vol. 40 ›› Issue (7): 207-215.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0152
收稿日期:2024-02-11
出版日期:2024-07-26
发布日期:2024-05-24
通讯作者:
云岚,女,博士,教授,研究方向:牧草种质资源与遗传育种;E-mail: yunlan@imau.edu.cn;作者简介:任晓敏,女,硕士研究生,研究方向:草种质资源与育种;E-mail: 2937923090@qq.com
基金资助:
REN Xiao-min1( ), YUN Lan1,2(
), YUN Lan1,2( ), AI Qian1, ZHAO Qiao3(
), AI Qian1, ZHAO Qiao3( )
)
Received:2024-02-11
Published:2024-07-26
Online:2024-05-24
摘要:
【目的】 异戊烯基转移酶(isopentenyl transferases, IPT)参与反式玉米素(trans-zeatin, tZ)的生物合成。tZ是调节植物生长发育和抵抗逆境胁迫的一种细胞分裂素类型,在促进芽生长和调控植物侧枝发育方面具有重要作用。探索IPT的功能,为后续新麦草产量性状改良提供了理论依据。【方法】 通过对新麦草IPT进行系统发育和多序列比对,探究其编码氨基酸序列的特征和同源性。通过花粉管通道法获得过表达PjIPT拟南芥(Arabidopsis thaliana)植株,并运用逆转录PCR(RT-PCR)和蛋白质印迹(Western blot, WB)对转基因植株进行验证,利用实时荧光定量逆转录PCR(RT-qPCR)对其组织表达模式进行分析,使用液质联用系统测定反式玉米素。【结果】 IPT在新麦草(Psathyrostachys juncea)和二粒小麦(Triticum dicoccoides)等麦类作物中具有高度同源性,且AtIPT9是PjIPT的同源蛋白。通过花粉管通道法获得过表达PjIPT拟南芥(Arabidopsis thaliana)植株,证明PjIPT上调植株分枝数。RT-PCR和蛋白质印迹分析显示,PjIPT在转录水平和蛋白水平均能够正常表达。利用不同部位组织RT-qPCR分析PjIPT的空间特异性表达,结果显示,PjIPT在拟南芥的分蘖节和叶中特异性表达。此外,反式玉米素的定量分析表明PjIPT通过参与tZ的生物合成去调控拟南芥植株分枝数。【结论】 PjIPT是上调植株分枝数的关键基因。
任晓敏, 云岚, 艾芊, 赵乔. 新麦草异戊烯基转移酶PjIPT基因的功能验证[J]. 生物技术通报, 2024, 40(7): 207-215.
REN Xiao-min, YUN Lan, AI Qian, ZHAO Qiao. Functional Verification of Isopentenyl Transferases PjIPT Gene in Psathyrostachys juncea[J]. Biotechnology Bulletin, 2024, 40(7): 207-215.
| 名称 Name | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| RT-qPCR | GCAAGCCATGGAGTACCTGT | TCTCATTGCGGAACCAGGTC |
| Actin | AGTGGTCGTACAACCGGTATTGT | GAGGAAGAGCATACCCCTCGTA |
| RT-IPT | CTTTCGTGAGAGTGGGCAGT | GAAGCGTCAACCCACTGGTA |
表1 引物列表
Table 1 List of primers
| 名称 Name | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| RT-qPCR | GCAAGCCATGGAGTACCTGT | TCTCATTGCGGAACCAGGTC |
| Actin | AGTGGTCGTACAACCGGTATTGT | GAGGAAGAGCATACCCCTCGTA |
| RT-IPT | CTTTCGTGAGAGTGGGCAGT | GAAGCGTCAACCCACTGGTA |
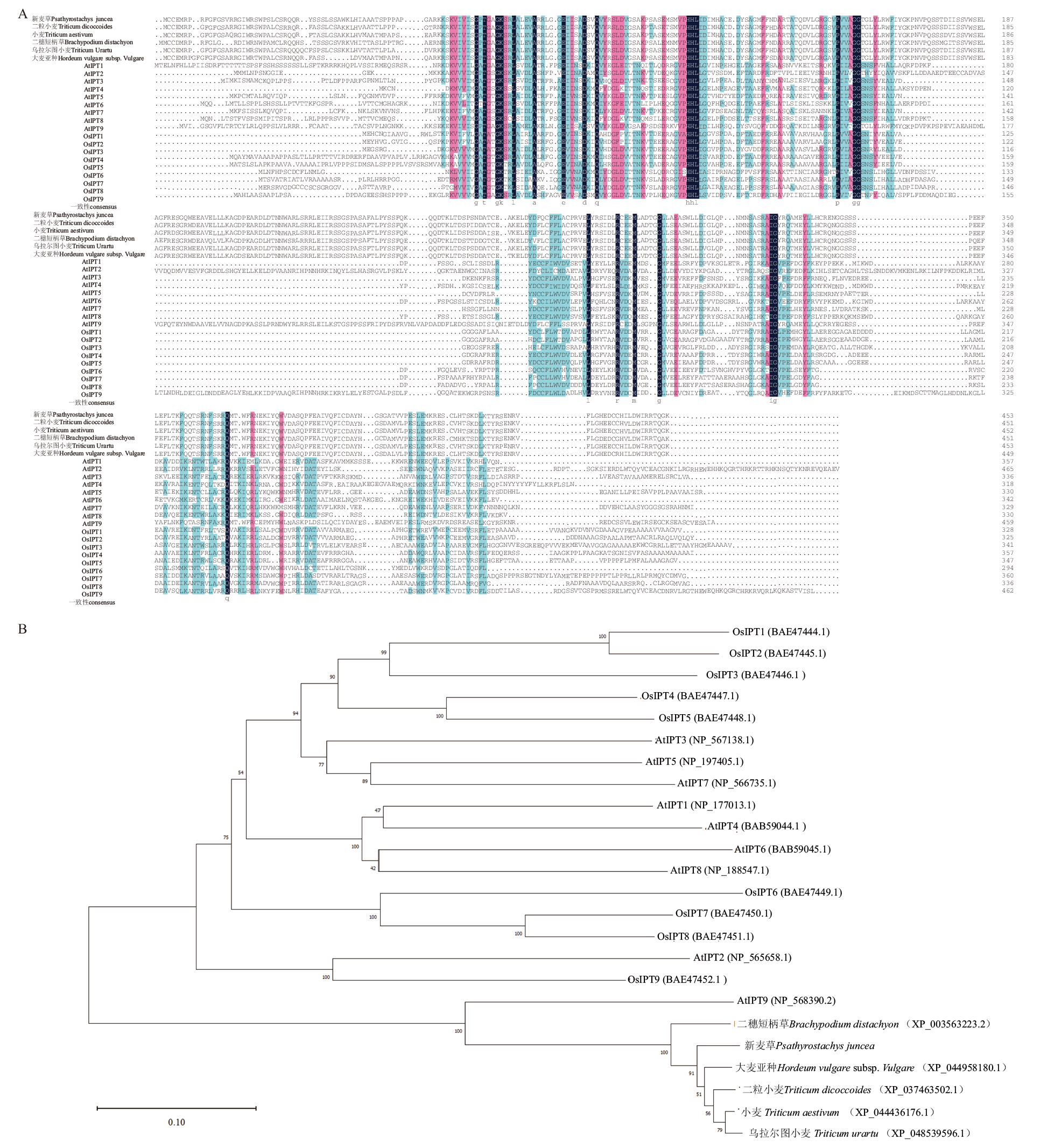
图2 基于IPT氨基酸序列的多序列比对及系统发育分析 A:不同物种IPT蛋白的多序列比对;B:不同物种IPT的系统进化树;Os:水稻;At:拟南芥
Fig. 2 Multiple sequence alignment and phylogenetic analysis based on IPT amino acid sequence A: Multiple sequence alignment of IPT proteins in different species. B: Phylogenetic tree of IPT in different species. Os: Oryza sativa; At: Arabidopsis thaliana
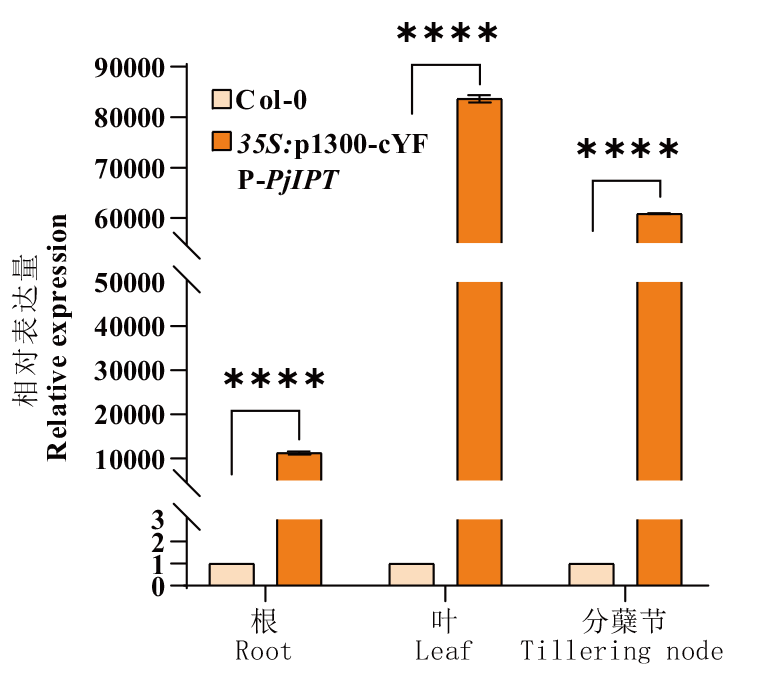
图4 PjIPT在拟南芥不同组织中的表达 ****表示表达水平差异极显著(P<0.000 1)
Fig. 4 Expression analysis of PjIPT in the different tissues of A. thaliana **** indicates significantly different(P<0.000 1)
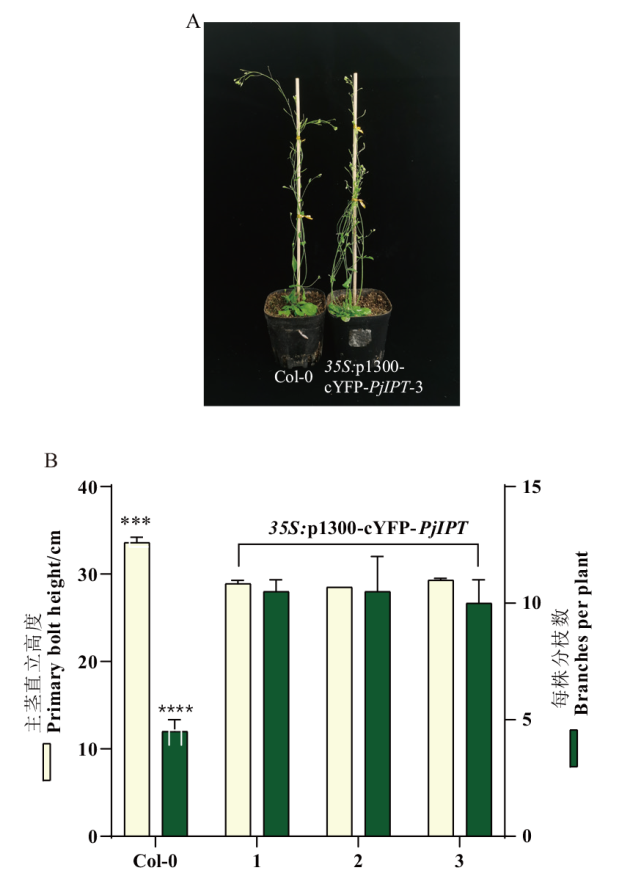
图5 35S:p1300-cYFP-PjIPT阳性植株表型分析 A:42日龄野生型与过表达PjIPT的代表性植株;B:42日龄Col-0和3个独立的35S:p1300-cYFP-PjIPT转基因拟南芥株系的分枝数和株高统计;***表示差异极显著(P<0.001)。下同
Fig. 5 Phenotypic analysis of 35S:p1300-cYFP-PjIPT positive plants A: Axillary branching of wild-type and overexpressed PjIPT in the representative plants described in B. B: Branch numbers and plant height in 42-day-old Col-0 and 3 independent 35S:p1300-cYFP-PjIPT transgenic Arabidopsis lines. ***: Extremely significant difference in(P <0.001). The same below
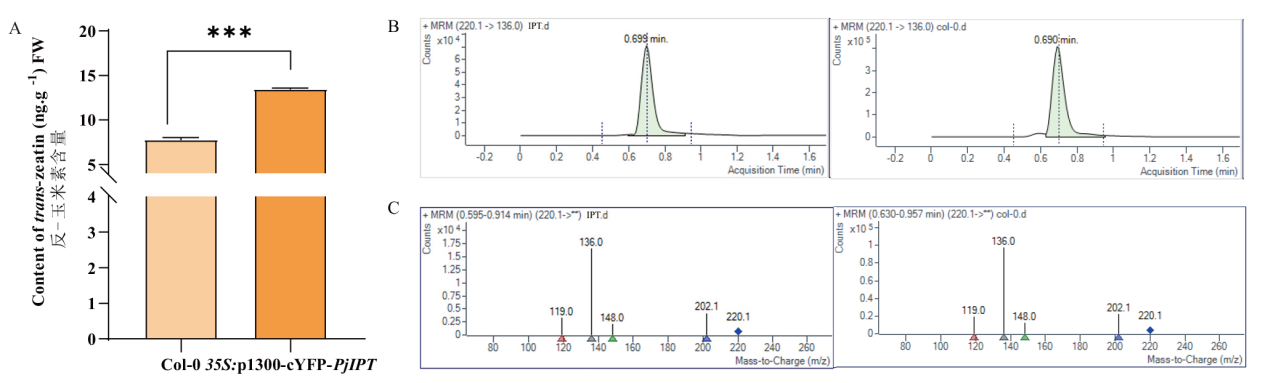
图6 反式玉米素定量分析 A:不同样品中反式玉米素含量的分析;B:MRM色谱图;C:MRM质谱图
Fig. 6 Quantitative analysis of trans-zeatin A: Analysis of trans-zeatin content in different samples; B: MRM chromatograms; C: MRM mass spectrographs
| [1] | 黄文, 李睿, 李卫军. 新麦草种子丸粒化包衣配方的筛选[J]. 现代农业科技, 2023(2): 178-183. |
| Huang W, Li R, Li WJ. Formula screening of pelleted and coated seeds of Psathyrostachys juncea[J]. Mod Agric Sci Technol, 2023(2): 178-183. | |
| [2] | 张晨, 云岚, 白亚利, 等. 新麦草种质资源的SSR遗传多样性分析[J]. 中国草地学报, 2017, 39(5): 24-30. |
| Zhang C, Yun L, Bai YL, et al. Genetic diversity analysis of 28 Psathyrostachys juncea germplasm resources based on SSR molecular markers[J]. Chin J Grassland, 2017, 39(5): 24-30. | |
| [3] | 张晴. 不同种源北沙参表型特征、香豆素类化合物含量及IPT基因表达差异研究[D]. 烟台: 烟台大学, 2023. |
| Zhang Q. Differences in phenotypic identificarion, coumarin content and expression of key enzyme genes in Glehnia littoralis from different origins[D]. Yantai: Yantai University, 2023. | |
| [4] | 周志湘. 蒺藜苜蓿(Medicago truncatula)细胞分裂素应答调控及IPT基因功能鉴定[D]. 北京: 北京林业大学, 2020. |
| Zhou ZX. Regulatory analysis of cytokinin response and function characterization of IPT genes in Medicago truncatula[D]. Beijing: Beijing Forestry University, 2020. | |
| [5] | 毕田田, 张骐, 代金玲, 等. 毛果杨PtIPT5基因的克隆及低温胁迫表达分析[J]. 江苏农业科学, 2023, 51(22): 14-23. |
| Bi TT, Zhang Q, Dai JL, et al. Cloning of PtIPT5 gene in Populus trichocarpa and its expression analysis under low temperature stress[J]. Jiangsu Agric Sci, 2023, 51(22): 14-23. | |
| [6] |
李皓, 张文, 赵旭勉, 等. 苹果异戊烯基转移酶基因家族(MdIPTs)的克隆与MdIPT5a功能分析[J]. 中国农业科学, 2011, 44(19): 4029-4036.
doi: 10.3864/j.issn.0578-1752.2011.19.013 |
| Li H, Zhang W, Zhao XM, et al. Molecular cloning of isopentenyl transferases genes family in Malus domestica borkh. and a preliminary functional analysis of MdIPT5a[J]. Sci Agric Sin, 2011, 44(19): 4029-4036. | |
| [7] | Zhang LP, Li M, Fu JY, et al. Genome-wide identification and expression analysis of Isopentenyl transferase family genes during development and resistance to abiotic stresses in tea plant(Camellia sinensis)[J]. Plants, 2022, 11(17): 2243. |
| [8] | Beznec A, Faccio P, Miralles DJ, et al. Stress-induced expression of IPT gene in transgenic wheat reduces grain yield penalty under drought[J]. J Genet Eng Biotechnol, 2021, 19(1): 67. |
| [9] | 阿不都热扎克·依沙克. 菊花侧枝发生相关基因CmERF053和CmIPT1的克隆与功能分析[D]. 北京: 中国农业大学, 2018. |
| Abdurazak I. Cloning and functional analysis of CmERFO53 and CmIPT1, which regulate the axillary bud developmentin chrysanthemum[D]. Beijing: China Agricultural University, 2018. | |
| [10] | 王珏琼. 异戊烯基转移酶(ipt)基因导入中间偃麦草及组织培养再生体系的建立[D]. 呼和浩特: 内蒙古大学, 2007. |
| Wang JQ. Genetic transformation of isopentenyl transferase gene to Thinopyrum intermedium and regeneration of transgenic plants[D]. Hohhot: Inner Mongolia University, 2007. | |
| [11] | 任晓敏, 云岚, 艾芊, 等. 新麦草IPT基因亚细胞定位、过表达载体构建及鉴定[J]. 西北植物学报, 2023, 43(9): 1441-1449. |
| Ren XM, Yun L, Ai Q, et al. Subcellular localization, overexpression vector construction and identification of IPT gene in Psathyrostachys juncea[J]. Acta Bot Boreali Occidentalia Sin, 2023, 43(9): 1441-1449. | |
| [12] |
Waters MT, Brewer PB, Bussell JD, et al. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones[J]. Plant Physiol, 2012, 159(3): 1073-1085.
doi: 10.1104/pp.112.196253 pmid: 22623516 |
| [13] | 李民吉. 桃ATP/ADP PpIPT基因鉴定与表达分析[D]. 泰安: 山东农业大学, 2015. |
| Li MJ. Identification and characterization of ATP/ADP PpIPT genes in peach[D]. Tai'an: Shandong Agricultural University, 2015. | |
| [14] | 王孝敬. 森林草莓IPT/CKX基因的进化及表达功能分析[D]. 南京: 南京农业大学, 2017. |
| Wang XJ. Studies on the evolution, expression patterns and functions of cytokinin biosynthesis and degradation genes in woodland strawberry[D]. Nanjing: Nanjing Agricultural University, 2017. | |
| [15] | Worakan P, Gujjar RS, Supaibulwatana K. Stable and reproducible expression of bacterial ipt gene under the control of SAM-specific promoter(pKNOX1)with interference of developmental patterns in transgenic Peperomia pellucida plants[J]. Front Plant Sci, 2022, 13: 984716. |
| [16] |
Takei K, Sakakibara H, Sugiyama T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana[J]. J Biol Chem, 2001, 276(28): 26405-26410.
doi: 10.1074/jbc.M102130200 pmid: 11313355 |
| [17] | Takei K, Yamaya T, Sakakibara H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin[J]. J Biol Chem, 2004, 279(40): 41866-41872. |
| [18] | Chen C, Sleper DA, Johal GS. Comparative RFLP mapping of meadow and tall fescue[J]. Theor Appl Genet, 1998, 97(1): 255-260. |
| [19] |
Miyawaki K, Matsumoto-Kitano M, Kakimoto T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate[J]. Plant J, 2004, 37(1): 128-138.
doi: 10.1046/j.1365-313x.2003.01945.x pmid: 14675438 |
| [20] |
Sakamoto T, Sakakibara H, Kojima M, et al. Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice[J]. Plant Physiol, 2006, 142(1): 54-62.
doi: 10.1104/pp.106.085811 pmid: 16861569 |
| [21] | Brugière N, Humbert S, Rizzo N, et al. A member of the maize isopentenyl transferase gene family, Zea mays isopentenyl transferase 2(ZmIPT2), encodes a cytokinin biosynthetic enzyme expressed during kernel development[J]. Plant Mol Biol, 2008, 67(3): 215-229. |
| [22] | Mi XN, Wang XJ, Wu H, et al. Characterization and expression analysis of cytokinin biosynthesis genes in Fragaria vesca[J]. Plant Growth Regul, 2017, 82(1): 139-149. |
| [23] | Le DT, Nishiyama R, Watanabe Y, et al. Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels[J]. PLoS One, 2012, 7(8): e42411. |
| [24] | Li Z, Yun L, Ren XM, et al. Analysis of controlling genes for tiller growth of Psathyrostachys juncea based on transcriptome sequencing technology[J]. BMC Plant Biol, 2022, 22(1): 456. |
| [25] | Yang HK, Li Y, Zhao JR, et al. Regulating the composition and secondary structure of wheat protein through canopy shading to improve dough performance and nutritional index[J]. Food Res Int, 2023, 173(Pt 2): 113399. |
| [26] | Garrido-Balam M, Chel-Guerrero L, Gallegos-Tintoré S, et al. Nutritional characterization of quality protein maize(QPM)(Zea mays L.) protein concentrates[J]. Food Humanity, 2023, 1: 1250-1255. |
| [27] | Liu D, Wu HL, Cui SY, et al. Comprehensive optimization of western blotting[J]. Gels, 2023, 9(8): 652. |
| [28] | Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4[J]. Nature, 1970, 227(5259): 680-685. |
| [29] | Yang JQ, Han FY, Yang L, et al. Identification of reference genes for RT-qPCR analysis in Gleditsia microphylla under abiotic stress and hormone treatment[J]. Genes, 2022, 13(7): 1227. |
| [30] | Liu JH, Yang C, Bai MZ, et al. Selection and validation of reference genes for RT-qPCR analysis of different organs at various development stages in Caragana intermedia[J]. Open Life Sci, 2022, 17(1): 1155-1164. |
| [31] | Asif S, Jan R, Kim N, et al. Halotolerant endophytic bacteria alleviate salinity stress in rice(Oryza sativa L.) by modulating ion content, endogenous hormones, the antioxidant system and gene expression[J]. BMC Plant Biol, 2023, 23(1): 494. |
| [32] | Qi F, Wang F, Xiaoyang CX, et al. Gene expression analysis of different organs and identification of AP2 transcription factors in flax(Linum usitatissimum L.)[J]. Plants, 2023, 12(18): 3260. |
| [33] | 刘艳洁, 赵晓冰, 王枫, 等. 细胞分裂素受体基因SlHK4突变对番茄开花时间的影响[J]. 南京农业大学学报, 2024, 47(3): 424-434. |
| Liu YJ, Zhao XB, Wang F, et al. Effects of cytokinin receptor gene SlHK4 mutation on flowering time of tomato[J]. J Nanjing Agric Univ, 2024, 47(3): 424-434. | |
| [34] | Kiba T, Mizutani K, Nakahara A, et al. The trans-Zeatin-type side-chain modification of cytokinins controls rice growth[J]. Plant Physiol, 2023, 192(3): 2457-2474. |
| [35] |
Kuroha T, Kato H, Asami T, et al. A trans-Zeatin riboside in root xylem sap negatively regulates adventitious root formation on cucumber hypocotyls[J]. J Exp Bot, 2002, 53(378): 2193-2200.
pmid: 12379786 |
| [36] | Antoniadi I, Mateo-Bonmatí E, Pernisová M, et al. IPT9, a cis-Zeatin cytokinin biosynthesis gene, promotes root growth[J]. Front Plant Sci, 2022, 13: 932008. |
| [37] | Eisermann I, Motyka V, Kümmel S, et al. CgIPT1 is required for synthesis of cis-Zeatin cytokinins and contributes to stress tolerance and virulence in Colletotrichum graminicola[J]. Fungal Genet Biol, 2020, 143: 103436. |
| [1] | 杜泽光, 任少文, 张凤勤, 李梅兰, 李改珍, 齐仙惠. 大白菜BrMLP328的克隆、表达及功能验证[J]. 生物技术通报, 2024, 40(4): 122-129. |
| [2] | 杨伟成, 孙岩, 杨倩, 王壮琳, 马菊花, 薛金爱, 李润植. 陆地棉FAX家族的全基因组鉴定及GhFAX1的功能分析[J]. 生物技术通报, 2024, 40(3): 155-169. |
| [3] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [4] | 陈妤, 朱沛煌, 李荣, 朱灵芝, 季孔庶. 植物异戊烯基转移酶研究进展[J]. 生物技术通报, 2021, 37(2): 149-161. |
| [5] | 乔孟欣, 李素贞, 陈景堂. 玉米铁还原酶基因ZmFRO2的功能分析[J]. 生物技术通报, 2020, 36(11): 9-20. |
| [6] | 刘丹, 吴凤芝. 分蘖洋葱查尔酮异构酶基因的克隆及表达载体的构建[J]. 生物技术通报, 2014, 30(10): 88-93. |
| [7] | 董绍旺;王爱英;孙建富;沈海涛;祝建波;. 转P_(rd29A)-ipt基因对烟草根及侧芽生长的影响[J]. , 2010, 0(08): 137-140. |
| [8] | 张怡;李成伟;. 酵母异源功能互补在植物基因克隆中的应用[J]. , 2010, 0(07): 14-21. |
| [9] | . 文摘[J]. , 2002, 0(03): 56-57. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||