








生物技术通报 ›› 2024, Vol. 40 ›› Issue (4): 122-129.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1082
杜泽光( ), 任少文, 张凤勤, 李梅兰, 李改珍, 齐仙惠(
), 任少文, 张凤勤, 李梅兰, 李改珍, 齐仙惠( )
)
收稿日期:2023-11-17
出版日期:2024-04-26
发布日期:2024-04-30
通讯作者:
齐仙惠,女,博士,副研究员,研究方向:园艺植物生物技术与遗传改良;E-mail: 651345642@qq.com作者简介:杜泽光,男,硕士研究生,研究方向:园艺植物生物技术与遗传改良;E-mail: 1527246146@qq.com
基金资助:
DU Ze-guang( ), REN Shao-wen, ZHANG Feng-qin, LI Mei-lan, LI Gai-zhen, QI Xian-hui(
), REN Shao-wen, ZHANG Feng-qin, LI Mei-lan, LI Gai-zhen, QI Xian-hui( )
)
Received:2023-11-17
Published:2024-04-26
Online:2024-04-30
摘要:
【目的】克隆大白菜BrMLP328基因,对其表达模式进行分析,并验证对花期调控的功能,为进一步探究该基因在大白菜成花调控过程的作用机理奠定基础。【方法】运用RT-PCR克隆BrMLP328,并进行生物信息学分析;利用RT-qPCR测定该基因的相对表达量;构建过表达载体并通过蘸花法转化野生型拟南芥,比较T2代植株与野生型的开花时间差异。【结果】BrMLP328的CDS全长为456 bp,编码151个氨基酸,蛋白相对分子质量为17 493.82 Da,定位于细胞核。BrMLP328在大白菜茎中的表达量最高,根中次之,花蕾中最低;茎尖生长点中的表达量表现为春化后升高,之后在花芽分化阶段迅速下降,并维持在很低的水平。过表达BrMLP328的拟南芥开花时间比野生型延迟了1.46-3.09 d。【结论】从大白菜中克隆得到BrMLP328基因,其表达量在不同组织及不同成花阶段有所不同,该基因能够延迟开花。
杜泽光, 任少文, 张凤勤, 李梅兰, 李改珍, 齐仙惠. 大白菜BrMLP328的克隆、表达及功能验证[J]. 生物技术通报, 2024, 40(4): 122-129.
DU Ze-guang, REN Shao-wen, ZHANG Feng-qin, LI Mei-lan, LI Gai-zhen, QI Xian-hui. Cloning,Expression and Functional Identification of BrMLP328 Gene in Brassica rapa subsp. pekinensis[J]. Biotechnology Bulletin, 2024, 40(4): 122-129.
| 用途 Purpose | 名称 Name | 序列 Sequence(5'-3') |
|---|---|---|
| 基因克隆 Gene cloning | 026653-1 | F:ATGGCGACGTCGGGAACA R:TTAAGCTTTGACGACATGATTGTCC |
| 大白菜RT-qPCR Fluorescent quantitative PCR in Chinese cabbage | ACTIN | F:CGGTGTCATGGTTGGGAGA R:CGTGCTCGATGGGGTACTTC |
| 026653-2 | F:CCACCACATCCAAGGTGTCA R:AACACCTCCGGTTTCCCATC | |
| 阳性鉴定 Positive identification | 026653-3 | F:GACGCACAATCCCACTATCC R:TCGGTTCGCTTCTCCCAGAT |
| 拟南芥RT-qPCR Fluorescent quantitative PCR in Arabidopsis thaliana | ACTII | F:CACACTGGAGTGATGGTTGG R:ATTGGCCTTGGGGTTAAGAG |
表1 试验中用到的引物序列
Table 1 Primer sequences used in the experiment
| 用途 Purpose | 名称 Name | 序列 Sequence(5'-3') |
|---|---|---|
| 基因克隆 Gene cloning | 026653-1 | F:ATGGCGACGTCGGGAACA R:TTAAGCTTTGACGACATGATTGTCC |
| 大白菜RT-qPCR Fluorescent quantitative PCR in Chinese cabbage | ACTIN | F:CGGTGTCATGGTTGGGAGA R:CGTGCTCGATGGGGTACTTC |
| 026653-2 | F:CCACCACATCCAAGGTGTCA R:AACACCTCCGGTTTCCCATC | |
| 阳性鉴定 Positive identification | 026653-3 | F:GACGCACAATCCCACTATCC R:TCGGTTCGCTTCTCCCAGAT |
| 拟南芥RT-qPCR Fluorescent quantitative PCR in Arabidopsis thaliana | ACTII | F:CACACTGGAGTGATGGTTGG R:ATTGGCCTTGGGGTTAAGAG |
| 软件名称 Software name | 软件网址 Software website |
|---|---|
| ProtParam | http://web.expasy.org/protparam/ |
| ProtScale | http://web.expasy.org/protscale/ |
| SOPMA | http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopm.html |
| SWISS-MODEL | http://swissmodel.expasy.org/ |
| Plant-mPLoc | http://www.csbio.sjtu.edu.cn/bioinf/plantmulti/ |
表2 本试验使用的主要在线软件
Table 2 Primary online software employed in the experiment
| 软件名称 Software name | 软件网址 Software website |
|---|---|
| ProtParam | http://web.expasy.org/protparam/ |
| ProtScale | http://web.expasy.org/protscale/ |
| SOPMA | http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopm.html |
| SWISS-MODEL | http://swissmodel.expasy.org/ |
| Plant-mPLoc | http://www.csbio.sjtu.edu.cn/bioinf/plantmulti/ |

图2 BrMLP328的生物信息学分析 A:BrMLP328的氨基酸序列多重比对;B:BrMLP328与其他同源蛋白的亲缘关系树;C:BrMLP328的亲疏水性分析;D:BrMLP328的二级结构预测;E:BrMLP328的三级结构预测
Fig. 2 Bioinformatics analysis of BrMLP328 A: Multiple alignment of BrMLP328 amino acid sequences. B: Phylogenetic tree of BrMLP328 homologous proteins. C: Analysis of hydrophilicity and hydrophobicity of BrMLP328. D: Prediction of secondary structures of BrMLP328 proteins. E: Prediction of tertiary structures of BrMLP328 proteins
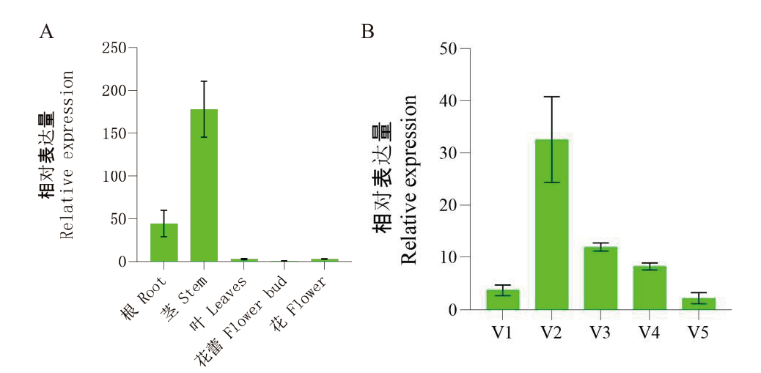
图3 大白菜不同器官(A)和成花不同阶段茎尖生长点(B)中BrMLP328的表达
Fig. 3 Expressions of BrMLP328 in different organs(A)and different developmental stages(B)of stem tip growth point
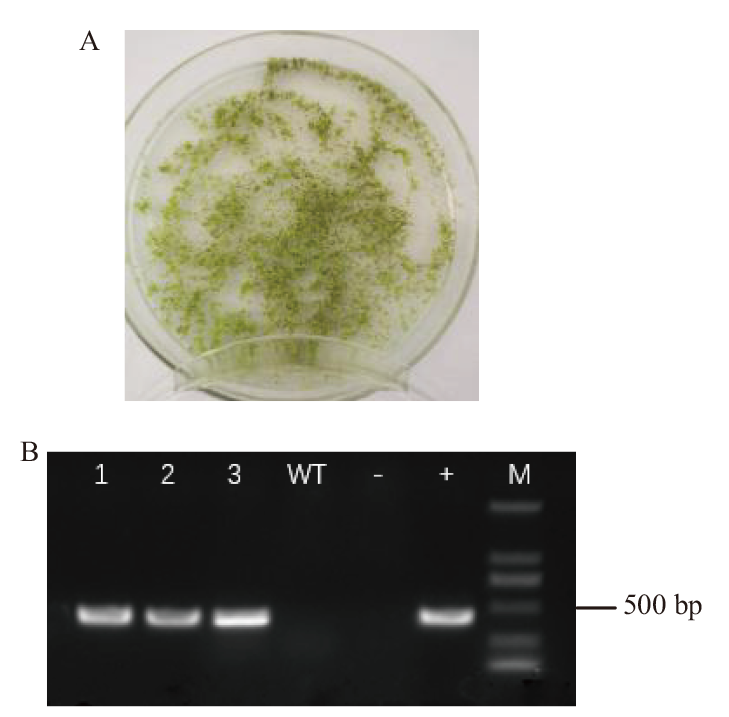
图4 T1代抗性植株的筛选及阳性鉴定 A:T1代抗性植株筛选;B:T1代抗性植株阳性鉴定(M:DL2000 marker,1-3:抗性植株,WT:野生型对照,-:阴性对照,+:阳性质粒对照)
Fig. 4 Screening and positive identification of T1 resistant plants A: Screening of resistant plants in T1 generation. B: Positive identification of resistant plants in T1 generation.(M: DL2000 marker. Label 1-3 is the number of resistant seedlings. “-” is the pure water blank control, “+” is the plasmid control and “WT” is the wild type plant control.)
| T1代植株编号 Number of T1 plant | T2代植株开花时间 Days of flowering after sowing of T2 plant/d | 比野生型植株延迟开花天数 Average number of days in flowering later than the wild type/d |
|---|---|---|
| WT | 21.23 ± 0.45 | - |
| Bra026653-1 | 24.32 ± 1.62 | 3.09 |
| Bra026653-2 | 22.69 ± 2.38 | 1.46 |
| Bra026653-3 | 23.26 ± 0.94 | 2.03 |
表3 T2代转基因植株与野生型的开花时间统计
Table 3 Statistics of flowering time between T2 transgenic plants and the wild type
| T1代植株编号 Number of T1 plant | T2代植株开花时间 Days of flowering after sowing of T2 plant/d | 比野生型植株延迟开花天数 Average number of days in flowering later than the wild type/d |
|---|---|---|
| WT | 21.23 ± 0.45 | - |
| Bra026653-1 | 24.32 ± 1.62 | 3.09 |
| Bra026653-2 | 22.69 ± 2.38 | 1.46 |
| Bra026653-3 | 23.26 ± 0.94 | 2.03 |
| [1] |
赖佳, 韦树谷, 黄玲, 等. 白菜类蔬菜种质资源抽薹性状鉴定评价[J]. 中国农学通报, 2022, 38(28): 41-47.
doi: 10.11924/j.issn.1000-6850.casb2022-0410 |
|
Lai J, Wei SG, Huang L, et al. Identification and evaluation on bolting traits of Chinese cabbage group germplasm resources[J]. Chin Agric Sci Bull, 2022, 38(28): 41-47.
doi: 10.11924/j.issn.1000-6850.casb2022-0410 |
|
| [2] |
Su TB, Wang WH, Li PR, et al. A genomic variation map provides insights into the genetic basis of spring Chinese cabbage(Brassica rapa ssp. pekinensis)selection[J]. Mol Plant, 2018, 11(11): 1360-1376.
doi: 10.1016/j.molp.2018.08.006 URL |
| [3] | 周麟笔, 马关鹏, 赵夏云, 等. 大白菜BrFLC及BrFRI-LIKE基因对低温的响应及其与抽薹性状的关系研究[J]. 种子, 2023, 42(4): 41-48. |
| Zhou LB, Ma GP, Zhao XY, et al. Response of BrFLC and BrFRI-LIKE genes to low temperature and their relationship with bolting trait in Chinese cabbage[J]. Seed, 2023, 42(4): 41-48. | |
| [4] | 罗睿, 郭建军. 植物开花时间: 自然变异与遗传分化[J]. 植物学报, 2010, 45(1): 109-118. |
| Luo R, Guo JJ. Plant flowering time: natural variation in the field and its role in determining genetic differentiation[J]. Chin Bull Bot, 2010, 45(1): 109-118. | |
| [5] | Li X, Shen CW, Chen RX, et al. Function of BrSOC1b gene in flowering regulation of Chinese cabbage and its protein interaction[J]. Planta, 2023, 258(1): 21. |
| [6] |
Nessler CL, Burnett RJ. Organization of the major latex protein gene family in opium poppy[J]. Plant Mol Biol, 1992, 20(4): 749-752.
doi: 10.1007/BF00046460 pmid: 1450390 |
| [7] |
Nessler CL, Kurz WGW, Pelcher LE. Isolation and analysis of the major latex protein genes of opium poppy[J]. Plant Mol Biol, 1990, 15(6): 951-953.
pmid: 2103486 |
| [8] |
Pozueta-Romero J, Klein M, Houlné G, et al. Characterization of a family of genes encoding a fruit-specific wound-stimulated protein of bell pepper(Capsicum annuum): identification of a new family of transposable elements[J]. Plant Mol Biol, 1995, 28(6): 1011-1025.
pmid: 7548820 |
| [9] |
Chen JY, Dai XF. Cloning and characterization of the Gossypium hirsutum major latex protein gene and functional analysis in Arabidopsis thaliana[J]. Planta, 2010, 231(4): 861-873.
doi: 10.1007/s00425-009-1092-2 URL |
| [10] | 李瑶, 王雷, 吴琼, 等. 植物乳胶蛋白研究进展[J]. 现代农业科技, 2021(19): 202-204. |
| Li Y, Wang L, Wu Q, et al. Research progress of plant latex protein[J]. Mod Agric Sci Technol, 2021(19): 202-204. | |
| [11] | 王雷, 孙尧, 李瑶, 等. 植物乳胶蛋白研究进展[J]. 安徽农业科学, 2021, 49(10): 12-14. |
| Wang L, Sun Y, Li Y, et al. Research progress of plant major latex protein[J]. J Anhui Agric Sci, 2021, 49(10): 12-14. | |
| [12] | 刘倩莹. 甘蓝MLP基因家族分析与BoMLP423的功能研究[D]. 重庆: 西南大学, 2022. |
| Liu QY. Analysis of MLP family genes and functional research of BoMLP423 in Brassica oleracea[D]. Chongqing: Southwest University, 2022. | |
| [13] |
Guo D, Wong WS, Xu WZ, et al. Cis-cinnamic acid-enhanced 1 gene plays a role in regulation of Arabidopsis bolting[J]. Plant Mol Biol, 2011, 75(4-5): 481-495.
doi: 10.1007/s11103-011-9746-4 pmid: 21298397 |
| [14] |
Phee BK, Park S, Cho JH, et al. Comparative proteomic analysis of blue light signaling components in the Arabidopsis cryptochrome 1 mutant[J]. Mol Cells, 2007, 23(2): 154-160.
doi: 10.1016/S1016-8478(23)07368-5 URL |
| [15] |
Mockler TC, Yu XH, Shalitin D, et al. Regulation of flowering time in Arabidopsis by K homology domain proteins[J]. Proc Natl Acad Sci USA, 2004, 101(34): 12759-12764.
doi: 10.1073/pnas.0404552101 pmid: 15310842 |
| [16] |
Osmark P, Boyle B, Brisson N. Sequential and structural homology between intracellular pathogenesis-related proteins and a group of latex proteins[J]. Plant Mol Biol, 1998, 38(6): 1243-1246.
pmid: 9869429 |
| [17] | Zhu LM, Ni WC, Liu S, et al. Transcriptomics analysis of apple leaves in response to Alternaria alternata apple pathotype infection[J]. Front Plant Sci, 2017, 8: 22. |
| [18] |
Ruperti B, Bonghi C, Ziliotto F, et al. Characterization of a major latex protein(MLP)gene down-regulated by ethylene during peach fruitlet abscission[J]. Plant Sci, 2002, 163(2): 265-272.
doi: 10.1016/S0168-9452(02)00094-8 URL |
| [19] |
Shang MY, Wang XT, Zhang J, et al. Genetic regulation of GA metabolism during vernalization, floral bud initiation and development in pak choi(Brassica rapa ssp. chinensis makino)[J]. Front Plant Sci, 2017, 8: 1533.
doi: 10.3389/fpls.2017.01533 URL |
| [20] | 宋红霞. 普通白菜成花转变基因表达谱比较及BrcLFY基因克隆与表达分析[D]. 晋中: 山西农业大学, 2016. |
| Song HX. Comparison of floral transition gene expression profiles and cloning and expression analysis of BrcLFY gene in Chinese cabbage[D]. Jinzhong: Shanxi Agricultural University, 2016. | |
| [21] |
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice[J]. Nucleic Acids Res, 1994, 22(22): 4673-4680.
doi: 10.1093/nar/22.22.4673 pmid: 7984417 |
| [22] |
Sarwar R, Geng R, Li L, et al. Genome-wide prediction, functional divergence, and characterization of stress-responsive BZR transcription factors in B. napus[J]. Front Plant Sci, 2022, 12: 790655.
doi: 10.3389/fpls.2021.790655 URL |
| [23] | 孙菲菲, 蒋芳玲, 侯喜林, 等. 白菜亚硝酸还原酶基因BcNiR的克隆及表达分析[J]. 园艺学报, 2009, 36(10): 1511-1518. |
| Sun FF, Jiang FL, Hou XL, et al. Molecular cloning and characterization of nitrite reductase gene BcNiR from non-heading Chinese cabbage[J]. Acta Hortic Sin, 2009, 36(10): 1511-1518. | |
| [24] |
Yuan GP, He SS, Bian SX, et al. Genome-wide identification and expression analysis of major latex protein(MLP)family genes in the apple(Malus domestica Borkh.) genome[J]. Gene, 2020, 733: 144275.
doi: 10.1016/j.gene.2019.144275 URL |
| [25] |
Goto J, Iwabuchi A, Yoshihara R, et al. Uptake mechanisms of polychlorinated biphenyls in Cucurbita pepo via xylem sap containing major latex-like proteins[J]. Environ Exp Bot, 2019, 162: 399-405.
doi: 10.1016/j.envexpbot.2019.03.019 URL |
| [26] | He SS, Yuan GP, Bian SX, et al. Major latex protein MdMLP423 negatively regulates defense against fungal infections in apple[J]. Int J Mol Sci, 2020, 21(5): 1879. |
| [27] |
Inui H, Sawada M, Goto J, et al. A major latex-like protein is a key factor in crop contamination by persistent organic pollutants[J]. Plant Physiol, 2013, 161(4): 2128-2135.
doi: 10.1104/pp.112.213645 pmid: 23404917 |
| [1] | 刘换换, 杨立春, 李火根. 北美鹅掌楸LtMYB305基因的克隆及功能分析[J]. 生物技术通报, 2024, 40(4): 179-188. |
| [2] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [3] | 杨伟成, 孙岩, 杨倩, 王壮琳, 马菊花, 薛金爱, 李润植. 陆地棉FAX家族的全基因组鉴定及GhFAX1的功能分析[J]. 生物技术通报, 2024, 40(3): 155-169. |
| [4] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [5] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| [6] | 朱毅, 柳唐镜, 宫国义, 张洁, 王晋芳, 张海英. 西瓜ClPP2C3克隆及表达分析[J]. 生物技术通报, 2024, 40(1): 243-249. |
| [7] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [8] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [9] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [10] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [11] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [12] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [13] | 侯瑞泽, 鲍悦, 陈启亮, 毛桂玲, 韦博霖, 侯雷平, 李梅兰. 普通白菜PRR5的克隆、表达及功能验证[J]. 生物技术通报, 2023, 39(10): 128-135. |
| [14] | 杨敏, 龙雨青, 曾娟, 曾梅, 周新茹, 王玲, 付学森, 周日宝, 刘湘丹. 灰毡毛忍冬UGTPg17、UGTPg36基因克隆及功能研究[J]. 生物技术通报, 2023, 39(10): 256-267. |
| [15] | 李秀青, 胡子曜, 雷建峰, 代培红, 刘超, 邓嘉辉, 刘敏, 孙玲, 刘晓东, 李月. 棉花黄萎病抗性相关基因GhTIFY9的克隆与功能分析[J]. 生物技术通报, 2022, 38(8): 127-134. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||