








生物技术通报 ›› 2024, Vol. 40 ›› Issue (7): 108-116.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0174
毋舒宁1,2( ), 苏永平1,2, 李冬雪3, 柏映国3, 刘波2(
), 苏永平1,2, 李冬雪3, 柏映国3, 刘波2( ), 张志伟1(
), 张志伟1( )
)
收稿日期:2024-02-21
出版日期:2024-07-26
发布日期:2024-07-30
通讯作者:
刘波,男,博士,研究员,研究方向:合成生物学;E-mail: lfb2500@163.com;作者简介:毋舒宁,女,硕士研究生,研究方向:林业有害生物控制与资源利用;E-mail: wsn06012023@163.com
基金资助:
WU Shu-ning1,2( ), SU Yong-ping1,2, LI Dong-xue3, BAI Ying-guo3, LIU Bo2(
), SU Yong-ping1,2, LI Dong-xue3, BAI Ying-guo3, LIU Bo2( ), ZHANG Zhi-wei1(
), ZHANG Zhi-wei1( )
)
Received:2024-02-21
Published:2024-07-26
Online:2024-07-30
摘要:
【目的】 谷氨酸棒杆菌是一种重要的工业微生物,挖掘可调控性元件可有效拓展谷氨酸棒杆菌研究和应用的广度和深度。【方法】 在谷氨酸棒杆菌组成型强启动子PH10上嵌入阻遏蛋白CymR结合的操纵序列CuO,构建了4-异丙基苯甲酸(Cumate)为诱导剂的启动子PH10-CuO。【结果】 绿色荧光蛋白作为报告基因的实验结果显示,无诱导剂时,谷氨酸棒杆菌工程菌相对荧光强度较低;以25 μg/mL 4-异丙基苯甲酸诱导12 h时,谷氨酸棒杆菌工程菌荧光强度达62 000,证明PH10-CuO具有很好的严谨性和诱导表达强度。构建了以启动子PH10-CuO调控recET和Cas12a表达的谷氨酸棒杆菌基因编辑质粒,实现谷氨酸棒杆菌染色体上靶标基因的精准编辑和外源基因插入。【结论】 诱导型启动子PH10-CuO具有表达强度高且渗漏表达低的特点,可用于谷氨酸棒杆菌中被其调控基因的时序性表达。
毋舒宁, 苏永平, 李冬雪, 柏映国, 刘波, 张志伟. 一种谷氨酸棒杆菌4-异丙基苯甲酸诱导型启动子的设计与应用[J]. 生物技术通报, 2024, 40(7): 108-116.
WU Shu-ning, SU Yong-ping, LI Dong-xue, BAI Ying-guo, LIU Bo, ZHANG Zhi-wei. Design and Application of a Cumate-inducible Promoter for Corynebacterium glutamicum[J]. Biotechnology Bulletin, 2024, 40(7): 108-116.
| 引物名称 Primer name | 引物序列 Primer sequence(5' -3') | 引物用途 Primer usage |
|---|---|---|
| cymR-F | CCAGGGTGGTTTTTCTTTCTATCGCTTGAACTTAGCGTAGC | 扩增CymR |
| cymR-R | AGAGTCAATTCAGGGTGGTGAATATGAGCCCGAAGCGCCGCAC | |
| LacIq-F | ATTCACCACCCTGAATTGACTCT | 扩增PlacIq |
| LacIq-R | GGATCAGCTTGCAATTCGCGCGCGAAGGCGAAGCGGCATTTACG | |
| Mazf-F | CACGGCGAAAGGATACTCATGGTAAGCCGATACGTACCCG | 扩增mazf |
| Mazf-R | GTCGACCTGCAGGCATGCCTACCCAATCAGTACGTTAATTTTGG | |
| gfp-F gfp-R | GCATGCCTGCAGGTCGACATGGTGAGCAAGGGCGAGGA ACCCGGGGATCCTCTAGATTACTTGTACAGCTCGTCCATG | 扩增gfp |
| cymR/H10CuO-F cymR/H10CuO-R | ATGGGTCTTGTTGTTGGCAGACCGTATCCAAAGCATCCG AGGAAGAGTGGTTTTGTGCTCATGAGTATCCTTTCGCCGTGCT | 扩增cymR/H10CuO |
| recET-F | ATGAGCACAAAACCACTCTTCCT | 扩增recET |
| recET-R | CTTCCTGGCATCTTCCAGCAAAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAATTATTCCTCTGAATTATCGATTACACTG | |
| H10CuO/cpf1-F | GTTCGCGGAATCATGACCGCTCAACCCTTACCGGTCG | PH10-CuO扩增 |
| H10CuO/cpf1-R | TCACAAACTCTTGGTAGATGGACATGAGTATCCTTTCGCCGTGCTC | |
| LysE-LF | TCAGTGGAACGAAAACTCACTGCGCGAGCAAGGAGAGTAC | 扩增lysE左同源臂 |
| LysE-LR | CGCCATGACAAACAAGGTGG | |
| LysE-RF | GTGGCACCGAGTCGGTGCTTTTTTTGAGGTAAGCGATGCCACCCCA | 扩增lysE右同源臂 |
| LysE-RR | TCTCATCCGCCAAAACAGCC GTTATGGTTTGCCGTCATGGCAGCATCTACAACAGTAGAAATTCGGATCCAT- TATACCTAGGACTGAGCTAGCTGTCAACCAAGAAGCTACCTCGTTGAACA | |
| asd/H10CuO-F | CCACCTTGTTTGTCATGGCGGCTCAACCCTTACCGGTCGGCTCTAAGCCGGCGGCGTATGGTAAGCTCTGTTATGTATAGTCCGAGCACGGCGAAAGGATACTC ATGACCACCATCGCAGTTGTT | 扩增asd |
| asd/H10CuO-R | AAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAATTACTTAACCAGCAGCTCAGCG |
表1 本研究所用引物
Table 1 Primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence(5' -3') | 引物用途 Primer usage |
|---|---|---|
| cymR-F | CCAGGGTGGTTTTTCTTTCTATCGCTTGAACTTAGCGTAGC | 扩增CymR |
| cymR-R | AGAGTCAATTCAGGGTGGTGAATATGAGCCCGAAGCGCCGCAC | |
| LacIq-F | ATTCACCACCCTGAATTGACTCT | 扩增PlacIq |
| LacIq-R | GGATCAGCTTGCAATTCGCGCGCGAAGGCGAAGCGGCATTTACG | |
| Mazf-F | CACGGCGAAAGGATACTCATGGTAAGCCGATACGTACCCG | 扩增mazf |
| Mazf-R | GTCGACCTGCAGGCATGCCTACCCAATCAGTACGTTAATTTTGG | |
| gfp-F gfp-R | GCATGCCTGCAGGTCGACATGGTGAGCAAGGGCGAGGA ACCCGGGGATCCTCTAGATTACTTGTACAGCTCGTCCATG | 扩增gfp |
| cymR/H10CuO-F cymR/H10CuO-R | ATGGGTCTTGTTGTTGGCAGACCGTATCCAAAGCATCCG AGGAAGAGTGGTTTTGTGCTCATGAGTATCCTTTCGCCGTGCT | 扩增cymR/H10CuO |
| recET-F | ATGAGCACAAAACCACTCTTCCT | 扩增recET |
| recET-R | CTTCCTGGCATCTTCCAGCAAAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAATTATTCCTCTGAATTATCGATTACACTG | |
| H10CuO/cpf1-F | GTTCGCGGAATCATGACCGCTCAACCCTTACCGGTCG | PH10-CuO扩增 |
| H10CuO/cpf1-R | TCACAAACTCTTGGTAGATGGACATGAGTATCCTTTCGCCGTGCTC | |
| LysE-LF | TCAGTGGAACGAAAACTCACTGCGCGAGCAAGGAGAGTAC | 扩增lysE左同源臂 |
| LysE-LR | CGCCATGACAAACAAGGTGG | |
| LysE-RF | GTGGCACCGAGTCGGTGCTTTTTTTGAGGTAAGCGATGCCACCCCA | 扩增lysE右同源臂 |
| LysE-RR | TCTCATCCGCCAAAACAGCC GTTATGGTTTGCCGTCATGGCAGCATCTACAACAGTAGAAATTCGGATCCAT- TATACCTAGGACTGAGCTAGCTGTCAACCAAGAAGCTACCTCGTTGAACA | |
| asd/H10CuO-F | CCACCTTGTTTGTCATGGCGGCTCAACCCTTACCGGTCGGCTCTAAGCCGGCGGCGTATGGTAAGCTCTGTTATGTATAGTCCGAGCACGGCGAAAGGATACTC ATGACCACCATCGCAGTTGTT | 扩增asd |
| asd/H10CuO-R | AAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAATTACTTAACCAGCAGCTCAGCG |
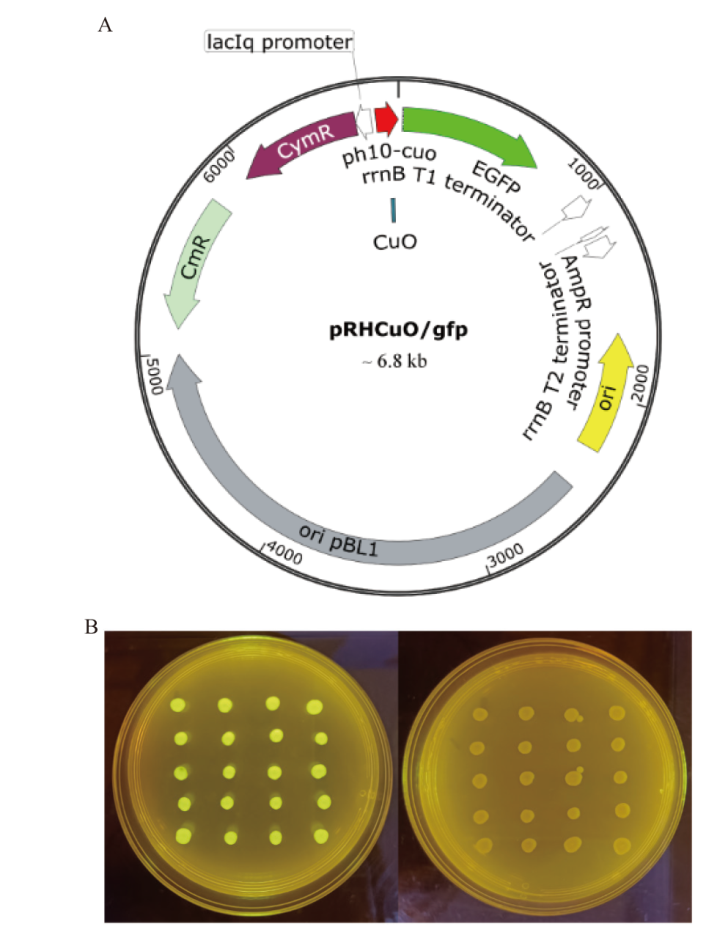
图3 启动子PH10-CuO性能鉴定 A:质粒pRHCuO/gfp;B:pRHCuO/gfp阳性转化子表征(左图:点接于含有4-异丙基苯甲酸的氯霉素抗性LB培养基,右图:点接于氯霉素抗性LB培养基)
Fig. 3 Characterization of PH10-CuO A: Plasmid pRHCuO/gfp. B: Profiles of positive transformants harboring pRHCuO/gfp on different media (Left: chloramphenicol-contained LB medium with 4-isopropylbenzoic acid; right: chloramphenicol-contained LB medium)
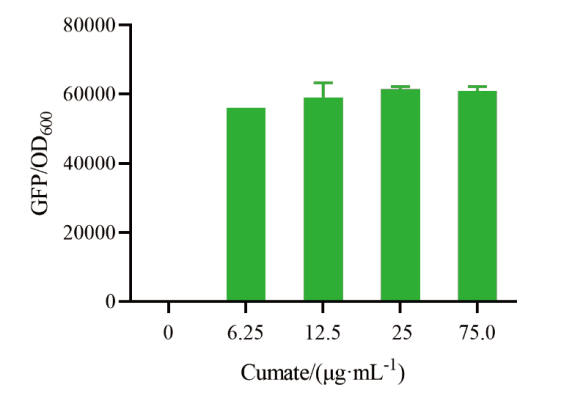
图4 启动子PH10-CuO性能鉴定 13032-01在不同4-异丙基苯甲酸浓度下诱导12 h时的相对荧光强度
Fig. 4 Characterization of PH10-CuO Fluorescence intensity at different 4-isopropylbenzoic acid concentrations at 12 h of 13032-01 induction
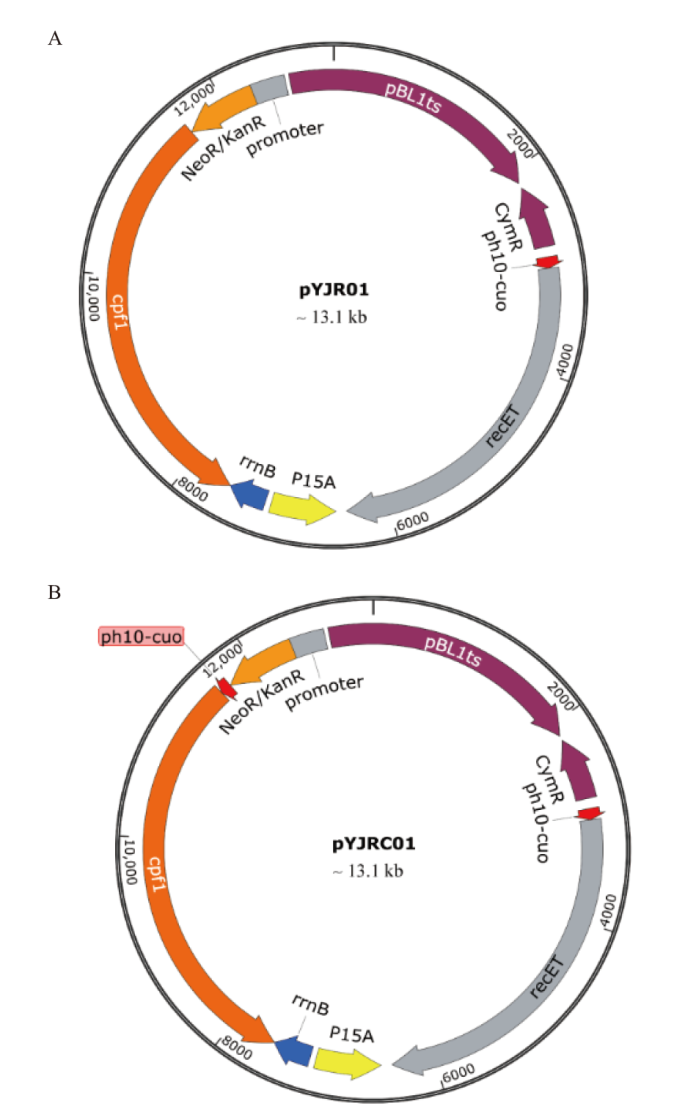
图5 基因编辑质粒 A:PH10-CuO调控recET表达的质粒pYJR01;B:PH10-CuO调控recET和cas12a表达的质粒pYJRC01
Fig. 5 Plasmids via gene editing A: PH10-CuO regulating the plasmid pYJR01 expressed with recET. B: PH10-CuO regulating the plasmid pYJR01 expressed with recET and cas12a
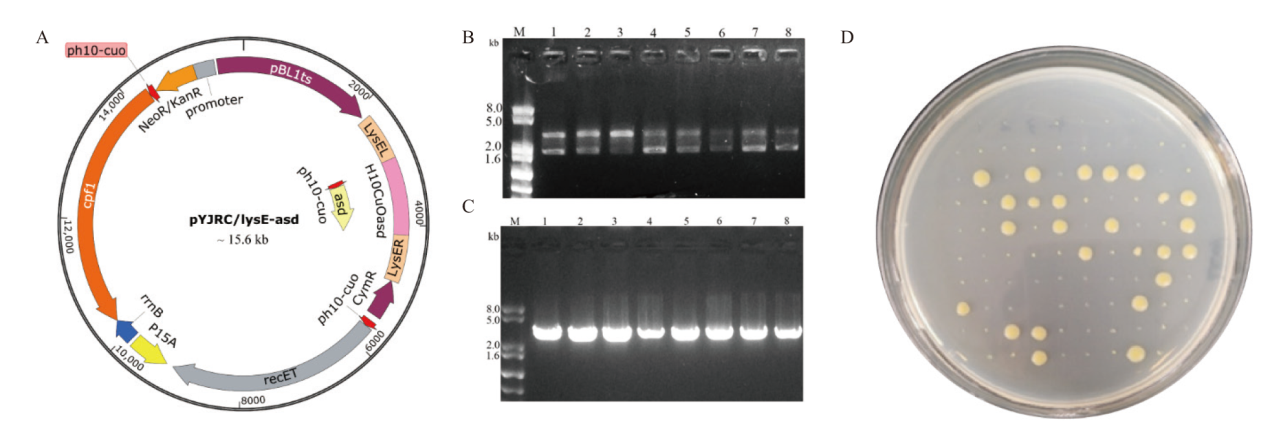
图6 基因编辑及质粒消除 A:用于敲除lysE和过表达asd的质粒pYJRC/lysE-asd;B:转化子验证(泳道M:DNA marker;泳道1-8:在含25 μg/mL 卡那霉素和12.5 μg/mL 4-异丙基苯甲酸的BHISG固体培养基上转化子染色体为模板时PCR扩增产物);C:转化子验证(泳道M:DNA marker;泳道1-8:在含25 μg/mL 卡那霉素和12.5 μg/mL 4-异丙基苯甲酸的液体培养基BHISG中再次培养24 h的菌体为模板时PCR扩增产物);D:质粒pYJRC/lysE-asd消除菌株筛选
Fig. 6 Gene editing and plasmid elimination A: Plasmid pYJRC/lysE-asd for deleting lysE and overexpressing asd. B: Transformants verification(Lane M: DNA marker; lane 1-8: PCR products with the chromosomal DNA of transformants on solid medium BHISG containing 25 μg/mL kanamycin and 12.5 μg/mL 4-isopropylbenzoic acid as templates. C: Transformants verification(Lane M: DNA marker; lane 1-8: PCR products with the chromosomal DNA of transformants cultured in liquid medium BHISG containing 25 μg/mL kanamycin and 12.5 μg/mL 4-isopropylbenzoic acid for 24 h as templates). D: Screen of transformants without plasmid pYJRC/lysE-asd
| [1] | Becker J, Rohles CM, Wittmann C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products[J]. Metab Eng, 2018, 50: 122-141. |
| [2] | Zhao NN, Qian L, Luo GJ, et al. Synthetic biology approaches to access renewable carbon source utilization in Corynebacterium glutamicum[J]. Appl Microbiol Biotechnol, 2018, 102(22): 9517-9529. |
| [3] |
Kalinowski J, Bathe B, Bartels D, et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins[J]. J Biotechnol, 2003, 104(1-3): 5-25.
doi: 10.1016/s0168-1656(03)00154-8 pmid: 12948626 |
| [4] |
Wendisch VF, Bott M, Eikmanns BJ. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids[J]. Curr Opin Microbiol, 2006, 9(3): 268-274.
pmid: 16617034 |
| [5] |
Blazeck J, Alper HS. Promoter engineering: recent advances in controlling transcription at the most fundamental level[J]. Biotechnol J, 2013, 8(1): 46-58.
doi: 10.1002/biot.201200120 pmid: 22890821 |
| [6] | Xu N, Wei L, Liu J. Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis[J]. World J Microbiol Biotechnol, 2019, 35(2): 33. |
| [7] | 刘莫识, 刘娇, 孙冠男, 等. 谷氨酸棒杆菌人工合成启动子文库的构建及应用[J]. 生物工程学报, 2022, 38(2): 831-842. |
| Liu MS, Liu J, Sun GN, et al. Construction and application of a synthetic promoter library for Corynebacterium glutamicum[J]. Chin J Biotechnol, 2022, 38(2): 831-842. | |
| [8] |
Eaton RW. P-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate[J]. J Bacteriol, 1997, 179(10): 3171-3180.
pmid: 9150211 |
| [9] | Choi YJ, Morel L, Le François T, et al. Novel, versatile, and tightly regulated expression system for Escherichia coli strains[J]. Appl Environ Microbiol, 2010, 76(15): 5058-5066. |
| [10] | Seo SO, Schmidt-Dannert C. Development of a synthetic cumate-inducible gene expression system for Bacillus[J]. Appl Microbiol Biotechnol, 2019, 103(1): 303-313. |
| [11] | Bai CX, van Wezel GP. CUBIC: a versatile cumate-based inducible CRISPRi system in Streptomyces[J]. ACS Synth Biol, 2023, 12(10): 3143-3147. |
| [12] | Pöschel L, Gehr E, Jordan P, et al. Expression of toxic genes in Methylorubrum extorquens with a tightly repressed, cumate-inducible promoter[J]. Antonie Van Leeuwenhoek, 2023, 116(12): 1285-1294. |
| [13] | Pan HQ, Shim A, Lubin MB, et al. Hopanoid lipids promote soybean- Bradyrhizobium symbiosis[J]. bioRxiv, 2023: 2023.09.04.556284. |
| [14] |
Mullick A, Xu Y, Warren R, et al. The cumate gene-switch: a system for regulated expression in mammalian cells[J]. BMC Biotechnol, 2006, 6: 43.
pmid: 17083727 |
| [15] | 张灵. 谷氨酸棒杆菌严格调控的启动子系统的建立[D]. 广州: 华南理工大学, 2021. |
| Zhang L. Establishment of tightly regulated promoter system in Corynebacterium glutamicum[D]. Guangzhou: South China University of Technology, 2021. | |
| [16] | Wei L, Xu N, Wang YR, et al. Promoter library-based module combination(PLMC)technology for optimization of threonine biosynthesis in Corynebacterium glutamicum[J]. Appl Microbiol Biotechnol, 2018, 102(9): 4117-4130. |
| [17] | Li Y, Ai YQ, Zhang JZ, et al. A novel expression vector for Corynebacterium glutamicum with an auxotrophy complementation system[J]. Plasmid, 2020, 107: 102476. |
| [18] | Zhang J, Yang FY, Yang YP, et al. Optimizing a CRISPR-Cpf1-based genome engineering system for Corynebacterium glutamicum[J]. Microb Cell Fact, 2019, 18(1): 60. |
| [19] | Su R, Wang T, Bo TD, et al. Enhanced production of D-pantothenic acid in Corynebacterium glutamicum using an efficient CRISPR-Cpf1 genome editing method[J]. Microb Cell Fact, 2023, 22(1):3. |
| [20] |
Kallscheuer N, Vogt M, Stenzel A, et al. Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and(2S)-flavanones[J]. Metab Eng, 2016, 38: 47-55.
doi: S1096-7176(16)30047-7 pmid: 27288926 |
| [21] |
上官玲玲, 卢慧芳, 夏会丽, 等. 谷氨酸棒杆菌细胞工厂构建与应用的研究进展[J]. 食品与发酵工业, 2022, 48(17): 313-320.
doi: 10.13995/j.cnki.11-1802/ts.030792 |
| Shangguan LL, Lu HF, Xia HL, et al. Research progress on construction and application of Corynebacterium glutamicum cell factory[J]. Food Ferment Ind, 2022, 48(17): 313-320. | |
| [22] | 龙梦飞, 徐美娟, 张显, 等. 合成生物学与代谢工程在谷氨酸棒杆菌产氨基酸中的应用[J]. 中国科学: 生命科学, 2019, 49(5): 541-552. |
| Long MF, Xu MJ, Zhang X, et al. Synthetic biology and metabolic engineering for amino acid production in Corynebacterium glutamicum[J]. Sci Sin Vitae, 2019, 49(5): 541-552. | |
| [23] | Wang B, Hu QT, Zhang Y, et al. A RecET-assisted CRISPR-Cas9 genome editing in Corynebacterium glutamicum[J]. Microb Cell Fact, 2018, 17(1): 63. |
| [24] | Deng C, Wu YK, Lv XQ, et al. Refactoring transcription factors for metabolic engineering[J]. Biotechnol Adv, 2022, 57: 107935. |
| [25] | Lu XY, Zhang MM, Li G, et al. Applications and research advances in the delivery of CRISPR/Cas9 systems for the treatment of inherited diseases[J]. Int J Mol Sci, 2023, 24(17): 13202. |
| [26] |
Liu X, Wu SR, Xu J, et al. Application of CRISPR/Cas9 in plant biology[J]. Acta Pharm Sin B, 2017, 7(3): 292-302.
doi: 10.1016/j.apsb.2017.01.002 pmid: 28589077 |
| [27] |
Tian PF, Wang J, Shen XL, et al. Fundamental CRISPR-Cas9 tools and current applications in microbial systems[J]. Synth Syst Biotechnol, 2017, 2(3): 219-225.
doi: 10.1016/j.synbio.2017.08.006 pmid: 29318202 |
| [28] |
Joung J, Konermann S, Gootenberg JS, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening[J]. Nat Protoc, 2017, 12(4): 828-863.
doi: 10.1038/nprot.2017.016 pmid: 28333914 |
| [29] | Liu J, Wang Y, Lu YJ, et al. Development of a CRISPR/Cas9 genome editing toolbox for Corynebacterium glutamicum[J]. Microb Cell Fact, 2017, 16(1): 205. |
| [30] | 王钰, 郑平, 孙际宾. 谷氨酸棒杆菌的代谢工程使能技术研究进展[J]. 生物工程学报, 2021, 37(5): 1603-1618. |
| Wang Y, Zheng P, Sun JB. Recent advances in developing enabling technologies for Corynebacterium glutamicum metabolic engineering[J]. Chin J Biotechnol, 2021, 37(5): 1603-1618. | |
| [31] |
Jiang Y, Qian FH, Yang JJ, et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum[J]. Nat Commun, 2017, 8: 15179.
doi: 10.1038/ncomms15179 pmid: 28469274 |
| [32] | 杨帆, 李寅. 新一代基因组编辑系统CRISPR/Cpf1[J]. 生物工程学报, 2017, 33(3): 361-371. |
| Yang F, Li Y. The new generation tool for CRISPR genome editing: CRISPR/Cpf1[J]. Chin J Biotechnol, 2017, 33(3): 361-371. | |
| [33] | 包心茹, 陈卯森, 钟洁, 等. CRISPR/Cas12a基因组编辑技术及应用[J]. 中国生物工程杂志, 2023, 43(10): 32-42. |
| Bao XR, Chen MS, Zhong J, et al. Characteristics and application of CRISPR/Cas12a genome editing technology[J]. China Biotechnol, 2023, 43(10): 32-42. | |
| [34] | Li N, Wang M, Yu SQ, et al. Optimization of CRISPR-Cas9 through promoter replacement and efficient production of L-homoserine in Corynebacterium glutamicum[J]. Biotechnol J, 2021, 16(8): e2100093. |
| [1] | 薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280. |
| [2] | 刘佳慧, 刘叶, 花尔并, 王猛. 谷氨酸棒杆菌中胞嘧啶碱基编辑工具的PAM拓展[J]. 生物技术通报, 2023, 39(9): 49-57. |
| [3] | 陈光, 李佳, 杜瑞英, 王旭. pOsHAK1:OsFLN2提高水稻的糖代谢水平和抗旱性[J]. 生物技术通报, 2022, 38(8): 92-100. |
| [4] | 聂立斌, 易铃欣, 邓妍, 盛琦, 吴晓玉, 张斌. 途径工程改造谷氨酸棒杆菌产莽草酸[J]. 生物技术通报, 2022, 38(6): 93-102. |
| [5] | 黄华媚, 白立宽, 刘叶, 李俊维, 王猛, 花尔并. BE3型胞嘧啶碱基编辑器在谷氨酸棒杆菌中的开发用[J]. 生物技术通报, 2020, 36(3): 95-101. |
| [6] | 聂志华, 朱蕾蕾. 生物素对发酵过程中MscCG外排L-谷氨酸的影响[J]. 生物技术通报, 2020, 36(10): 150-155. |
| [7] | 徐德雨, 郑小梅, 赵晶, 郑平, 赵树欣. 谷氨酸棒杆菌天冬氨酸激酶G359D突变解除赖氨酸与苏氨酸协同抑制的研究[J]. 生物技术通报, 2017, 33(11): 143-152. |
| [8] | 李田, 孙景宽, 刘京涛. 植物启动子研究进展[J]. 生物技术通报, 2015, 31(2): 18-25. |
| [9] | 余小霞, 田健, 刘晓青, 伍宁丰. 枯草芽孢杆菌表达系统及其启动子研究进展[J]. 生物技术通报, 2015, 31(2): 35-44. |
| [10] | 李濯雪,陈信波. 植物诱导型启动子及相关顺式作用元件研究进展[J]. 生物技术通报, 2015, 31(10): 8-15. |
| [11] | 石增秀 崔文璟 周丽 周哲敏. 谷氨酸棒杆菌L-天冬氨酸α-脱羧酶基因的克隆及重组酶性质研究[J]. 生物技术通报, 2013, 0(4): 110-115. |
| [12] | 罗玉常;窦文芳;张晓梅;史劲松;许正宏;. 谷氨酸棒杆菌ilvE基因的敲除对相关氨基酸合成的影响[J]. , 2012, 0(11): 185-191. |
| [13] | 马睿;陈华民;吴茂森;吴祖建;何晨阳;. 病原物诱导型植物启动子的研究方法[J]. , 2012, 0(11): 32-37. |
| [14] | 黄云雁;黎明;刘萌;孙昕;周丽颖;路福平;. 赖氨酸-尸胺反向转运蛋白cadB基因谷氨酸棒杆菌表达载体的构建及转化[J]. , 2012, 0(08): 94-100. |
| [15] | 伍展红;郑穗平;. 谷氨酸棒杆菌ldh基因的敲除[J]. , 2012, 0(02): 107-111. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||