








生物技术通报 ›› 2025, Vol. 41 ›› Issue (7): 164-171.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0150
王月琛1( ), 韩鑫骐1, 魏文敏1, 崔兆兰1, 罗阳美1, 陈鹏如1, 王海岗2, 刘龙龙2, 张莉1(
), 韩鑫骐1, 魏文敏1, 崔兆兰1, 罗阳美1, 陈鹏如1, 王海岗2, 刘龙龙2, 张莉1( ), 王纶2(
), 王纶2( )
)
收稿日期:2025-02-14
出版日期:2025-07-26
发布日期:2025-07-22
通讯作者:
王纶,男,研究员,研究方向 :糜子种质资源创新与利用;E-mail: wanglun976pzs@sina.com作者简介:王月琛,女,硕士研究生,研究方向 :作物遗传育种;E-mail: 2756309376@qq.com
基金资助:
WANG Yue-chen1( ), HAN Xin-qi1, WEI Wen-min1, CUI Zhao-lan1, LUO Yang-mei1, CHEN Peng-ru1, WANG Hai-gang2, LIU long-long2, ZHANG Li1(
), HAN Xin-qi1, WEI Wen-min1, CUI Zhao-lan1, LUO Yang-mei1, CHEN Peng-ru1, WANG Hai-gang2, LIU long-long2, ZHANG Li1( ), WANG Lun2(
), WANG Lun2( )
)
Received:2025-02-14
Published:2025-07-26
Online:2025-07-22
摘要:
目的 黍稷对我国旱作农业、盐碱地开发利用和救灾补种有着重要的作用,但是落粒是黍稷重要的产量限制因素。研究黍稷落粒的机制,对选育抗落粒的黍稷新品种,提高黍稷产量具有重要的意义。 方法 以落粒性强的‘野糜子’和落粒性弱的‘红粘糜’为材料,利用石蜡切片,研究不同落粒性的黍稷品种间离层细胞的结构差异。其次运用反向遗传学策略,鉴定与水稻落粒基因同源的黍稷落粒基因。 结果 首先,通过对‘野糜子’和‘红粘糜’的离区组织的电镜观察发现,‘野糜子’在开花后,形成明显的离层,而在‘红粘糜’中,没有观察到完整的离层。其次,对黍稷落粒基因在‘野糜子’小穗开花后1、20和35 d的表达情况进行分析发现,大部分基因在开花后1 d的表达量最高。最后,通过比较‘野糜子’和‘红粘糜’PmSh1-1基因的cDNA序列发现,‘红粘糜’PmSh1-1的cDNA比‘野糜子’的cDNA短,缺少第3个外显子。 结论 黍稷离层发育不完整,能够导致黍稷落粒性降低。水稻OsSh1的同源基因PmSh1-1可能是调控黍稷落粒的候选基因。
王月琛, 韩鑫骐, 魏文敏, 崔兆兰, 罗阳美, 陈鹏如, 王海岗, 刘龙龙, 张莉, 王纶. 黍稷落粒的生物学基础研究及落粒调控基因的鉴定[J]. 生物技术通报, 2025, 41(7): 164-171.
WANG Yue-chen, HAN Xin-qi, WEI Wen-min, CUI Zhao-lan, LUO Yang-mei, CHEN Peng-ru, WANG Hai-gang, LIU long-long, ZHANG Li, WANG Lun. Biological Basis Study for Grain Shattering in Proso Millet and Identification of Genes Regulating Grain Shattering[J]. Biotechnology Bulletin, 2025, 41(7): 164-171.
基因名称 Gene name | 基因编号 Gene ID | 编码蛋白 Protein name | 黍稷同源基因1 Homologous gene 1 | 黍稷同源基因2 Homologous gene 2 |
|---|---|---|---|---|
| qSH1 | Os01g0848400 | BEL1型同源异型蛋白 | PM13G13360 | PM07G28580 |
| OsSh1 | Os03g0650000 | YABBY转录因子 | PM01G33800 | PM02G06310 |
| sh4/SHA1 | Os04g0670900 | MYB转录因子 | PM16G23590 | PM15G25020 |
| SHAT1 | Os04g0649100 | AP2转录因子 | PM16G22000 | PM15G23430 |
| SH5 | Os05g0455200 | BELI型同源异型蛋白 | PM05G36060 | PM06G19790 |
| OsCPL1 | Os07g0207700 | CTD磷酸化酶 | PM15G01070 | PM16G00940 |
表1 水稻和黍稷落粒基因
Table 1 Grain shattering genes in rice (Oryza sativa) and proso millet (Panicum miliaceum)
基因名称 Gene name | 基因编号 Gene ID | 编码蛋白 Protein name | 黍稷同源基因1 Homologous gene 1 | 黍稷同源基因2 Homologous gene 2 |
|---|---|---|---|---|
| qSH1 | Os01g0848400 | BEL1型同源异型蛋白 | PM13G13360 | PM07G28580 |
| OsSh1 | Os03g0650000 | YABBY转录因子 | PM01G33800 | PM02G06310 |
| sh4/SHA1 | Os04g0670900 | MYB转录因子 | PM16G23590 | PM15G25020 |
| SHAT1 | Os04g0649100 | AP2转录因子 | PM16G22000 | PM15G23430 |
| SH5 | Os05g0455200 | BELI型同源异型蛋白 | PM05G36060 | PM06G19790 |
| OsCPL1 | Os07g0207700 | CTD磷酸化酶 | PM15G01070 | PM16G00940 |

图1 ‘野糜子’和‘红粘糜’的离区的组织学观察A:成熟‘野糜子’的小穗;B:‘野糜子’小穗石蜡切片;C:成熟‘红粘糜’的小穗;D:‘红粘糜’小穗石蜡切片
Fig. 1 Histological observation of the abscission zone of 'Yemizi' and 'Hongnianmi'A: Mature spikelet of 'Yemizi'. B: Paraffin section of 'Yemizi' spikelet. C: Mature spikelet of 'Hongnianmi'. D: Paraffin section of 'Hongnianmi' spikelet
基因名称 Gene name | 基因编号 Gene ID | ‘0390’染色体位置 Chromosome location in ‘0390’ | ‘晋黍7号’染色体位置 Chromosome location in ‘Jinsu7’ | ‘隆糜4号’染色体位置 Chromosome location in ‘Longmei4’ |
|---|---|---|---|---|
| PmSH5-1 | PM05G36060 | 05G | 3B | Chr03 |
| PmSH5-2 | PM06G19790 | 06G | 3A | Chr10 |
| PmqSH1-1 | PM07G28580 | 07G | 5B | Chr05 |
| PmqSH1-2 | PM13G13360 | 13G | 5A | Chr08 |
| PmSh1-1 | PM01G33800 | 01G | 9B | Chr01 |
| PmSh1-2 | PM02G06310 | 02G | 9A | Chr04 |
| PmCPL1-1 | PM15G01070 | 15G | 7B | Chr14 |
| PmCPL1-2 | PM16G00940 | 16G | 7A | Chr15 |
| PmSHAT1-1 | PM15G23430 | 15G | 7B | Chr14 |
| PmSHAT1-2 | PM16G22000 | 16G | 7A | Chr15 |
| Pmsh4/SHA1-1 | PM15G25020 | 15G | 7B | Chr14 |
| Pmsh4/SHA1-2 | PM16G23590 | 16G | 7A | Chr15 |
表2 黍稷落粒同源基因的染色体位置
Table 2 Chromosome location of homologous grain shattering genes in proso millet
基因名称 Gene name | 基因编号 Gene ID | ‘0390’染色体位置 Chromosome location in ‘0390’ | ‘晋黍7号’染色体位置 Chromosome location in ‘Jinsu7’ | ‘隆糜4号’染色体位置 Chromosome location in ‘Longmei4’ |
|---|---|---|---|---|
| PmSH5-1 | PM05G36060 | 05G | 3B | Chr03 |
| PmSH5-2 | PM06G19790 | 06G | 3A | Chr10 |
| PmqSH1-1 | PM07G28580 | 07G | 5B | Chr05 |
| PmqSH1-2 | PM13G13360 | 13G | 5A | Chr08 |
| PmSh1-1 | PM01G33800 | 01G | 9B | Chr01 |
| PmSh1-2 | PM02G06310 | 02G | 9A | Chr04 |
| PmCPL1-1 | PM15G01070 | 15G | 7B | Chr14 |
| PmCPL1-2 | PM16G00940 | 16G | 7A | Chr15 |
| PmSHAT1-1 | PM15G23430 | 15G | 7B | Chr14 |
| PmSHAT1-2 | PM16G22000 | 16G | 7A | Chr15 |
| Pmsh4/SHA1-1 | PM15G25020 | 15G | 7B | Chr14 |
| Pmsh4/SHA1-2 | PM16G23590 | 16G | 7A | Chr15 |
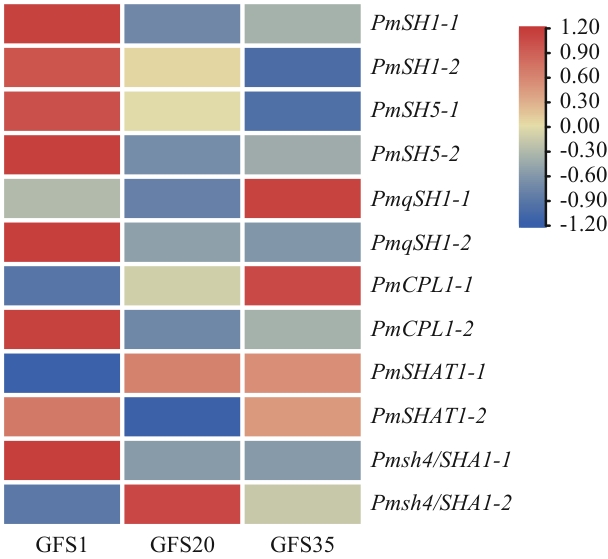
图4 黍稷落粒同源基因的组织特异性表达热图橘红色代表表达量上调,蓝色代表表达量下调,表达差异大于2倍视为显著;GFS1表示开花后1 d,GFS20表示开花后20 d,GFS35表示开花后35 d
Fig. 4 Heat-map showing the tissue-specific expressions of homologous grain shattering genes in proso milletOrange-red indicates up-regulated expression, blue indicates down-regulated, and expression difference greater than 2 times is considered significant. GFS1 refers to 1 d after flowering, GFS20 refers to 20 d after flowering, GFS35 refers to 35 d after flowering
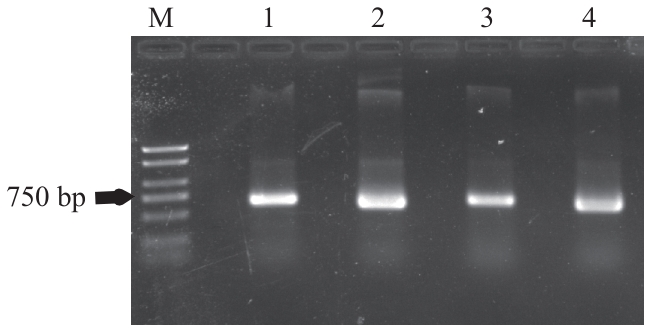
图5 PmSh1-1 cDNA的琼脂糖凝胶电泳检测图M:分子量标准;1和3:野糜子;2和4:红粘糜
Fig. 5 Detection of PmSh1-1 cDNA by agarose gel electrophoresisM: Marker; 1 and 3: Yemizi; 2 and 4: Hongnianmi

图6 PmSh1-1在‘野糜子’和‘红粘糜’中cDNA序列比对缺失的序列和点突变用红框标注
Fig. 6 cDNA sequence alignment of PmSh1-1 in 'Yemizi' and 'Hongnianmi'The indel sequence and SNPs are marked with red squares
| [1] | 王星玉, 王纶, 温琪汾. 黍稷的名实考证及规范 [J]. 植物遗传资源学报, 2010, 11(2): 132-138. |
| Wang XY, Wang L, Wen QF. Textual research and standardization of Chinese Name of broomcorn millet [J]. J Plant Genet Resour, 2010, 11(2): 132-138. | |
| [2] | Lu HY, Zhang JP, Liu KB, et al. Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10, 000 years ago [J]. Proc Natl Acad Sci USA, 2009, 106(18): 7367-7372. |
| [3] | 王纶, 王星玉. 中国黍稷种质资源研究 [M]. 北京: 中国农业科学技术出版社, 2018. |
| Wang L, Wang XY. Study on millet germplasm resources in China [M]. Beijing: China Agricultural Science and Technology Press, 2018. | |
| [4] | 刘敏轩, 许月, 陆平. 中国野生黍稷资源收集保存与遗传多样性研究进展 [J]. 植物遗传资源学报, 2020, 21(6): 1435-1445. |
| Liu MX, Xu Y, Lu P. Advances in germplasm collection and genetic diversity research of wild broomcorn millet in China [J]. J Plant Genet Resour, 2020, 21(6): 1435-1445. | |
| [5] | 苏占明, 李海, 皇甫红芳. 黍稷的营养成分与保健功效及种植建议 [J]. 种业导刊, 2020(4): 31-34. |
| Su ZM, Li H, Huangfu HF. Nutritional components, health care effects and planting suggestions of millet [J]. J Seed Ind Guide, 2020(4): 31-34. | |
| [6] | 李新月, 董燕, 李颖, 等. 五谷为养之黍米历史溯源与功能考证 [J]. 中国食品药品监管, 2022(8): 118-125. |
| Li XY, Dong Y, Li Y, et al. Historical traceability and functional research of broomcorn millet as a grain for raising [J]. China Food Drug Adm Mag, 2022(8): 118-125. | |
| [7] | 王纶, 王星玉, 温琪汾. 黍稷种质资源繁殖更新技术 [J]. 山西农业科学, 2012, 40(3): 227-232, 240. |
| Wang L, Wang XY, Wen QF. Regeneration and renew technical specification of broomcorn millet germplasm resources [J]. J Shanxi Agric Sci, 2012, 40(3): 227-232, 240. | |
| [8] | 谢文刚, 万依阳, 张宗瑜, 等. 禾本科植物落粒机理研究进展 [J]. 草业学报, 2021, 30(8): 186-198. |
| Xie WG, Wan YY, Zhang ZY, et al. Research progress on the seed-shattering mechanism of Poaceae plants [J]. Acta Prataculturae Sin, 2021, 30(8): 186-198. | |
| [9] | Maity A, Lamichaney A, Joshi DC, et al. Seed shattering: a trait of evolutionary importance in plants [J]. Front Plant Sci, 2021, 12: 657773. |
| [10] | Yu YQ, Hu H, Doust AN, et al. Divergent gene expression networks underlie morphological diversity of abscission zones in grasses [J]. New Phytol, 2020, 225(4): 1799-1815. |
| [11] | Yu YQ, Kellogg EA. Multifaceted mechanisms controlling grain disarticulation in the Poaceae [J]. Curr Opin Plant Biol, 2024, 81: 102564. |
| [12] | Parker TA, Lo S, Gepts P. Pod shattering in grain legumes: emerging genetic and environment-related patterns [J]. Plant Cell, 2021, 33(2): 179-199. |
| [13] | Yu YQ, Beyene G, Villmer J, et al. Grain shattering by cell death and fracture in Eragrostis tef [J]. Plant Physiol, 2023, 192(1): 222-239. |
| [14] | Ning J, He W, Wu LH, et al. The MYB transcription factor Seed Shattering 11 controls seed shattering by repressing lignin synthesis in African rice [J]. Plant Biotechnol J, 2023, 21(5): 931-942. |
| [15] | Wu H, He Q, He B, et al. Gibberellin signaling regulates lignin biosynthesis to modulate rice seed shattering [J]. Plant Cell, 2023, 35(12): 4383-4404. |
| [16] | Jiang LY, Ma X, Zhao SS, et al. The APETALA2-like transcription factor SUPERNUMERARY BRACT controls rice seed shattering and seed size [J]. Plant Cell, 2019, 31(1): 17-36. |
| [17] | Hofmann NR. SHAT1, A new player in seed shattering of rice [J]. Plant Cell, 2012, 24(3): 839. |
| [18] | Zhang YL, Xia QY, Jiang XQ, et al. Reducing seed shattering in weedy rice by editing SH4 and qSH1 genes: implications in environmental biosafety and weed control through transgene mitigation [J]. Biology, 2022, 11(12): 1823. |
| [19] | Wu H, He Q, Wang Q. Advances in rice seed shattering [J]. Int J Mol Sci, 2023, 24(10): 8889. |
| [20] | 吕树伟, 唐璇, 李晨. 水稻落粒性研究进展 [J]. 中国农业科学, 2025, 58(1): 1-9. |
| Lü SW, Tang X, Li C. Research progress on seed shattering of rice [J]. Sci Agric Sin, 2025, 58(1): 1-9. | |
| [21] | 王星玉, 王纶. 黍稷种质资源描述规范和数据标准 [M]. 北京: 中国农业出版社, 2006. |
| Wang XY, Wang L. Descriptors and data standard for broomcorn millet (Panicum miliaceum L.) [M]. Beijing: China Agriculture Press, 2006. | |
| [22] | Hodge JG, Kellogg EA. Abscission zone development in Setaria viridis and its domesticated relative, Setaria italica [J]. Am J Bot, 2016, 103(6): 998-1005. |
| [23] | Ishikawa R, Castillo CC, Htun TM, et al. A stepwise route to domesticate rice by controlling seed shattering and panicle shape [J]. Proc Natl Acad Sci USA, 2022, 119(26): e2121692119. |
| [24] | 张兰. 小麦落粒性与产量性状相关基因功能鉴定 [D]. 北京: 中国农业科学院, 2013. |
| Zhang L. Functional identification of genes related to grain dropping and yield traits in wheat [D]. Beijing: Chinese Academy of Agricultural Sciences, 2013. | |
| [25] | Liu HQ, Fang XJ, Zhou LN, et al. Transposon insertion drove the loss of natural seed shattering during foxtail millet domestication [J]. Mol Biol Evol, 2022, 39(6): msac078. |
| [26] | Lin ZW, Li XR, Shannon LM, et al. Parallel domestication of the Shattering1 genes in cereals [J]. Nat Genet, 2012, 44(6): 720-724. |
| [27] | Yu YQ, Hu H, Voytas DF, et al. The YABBY gene SHATTERING1 controls activation rather than patterning of the abscission zone in Setaria viridis [J]. New Phytol, 2023, 240(2): 846-862. |
| [28] | Ishikawa R. Genetic dissection of a reduced seed-shattering trait acquired in rice domestication [J]. Breed Sci, 2024, 74(4): 285-294. |
| [29] | Shi JP, Ma XX, Zhang JH, et al. Chromosome conformation capture resolved near complete genome assembly of broomcorn millet [J]. Nat Commun, 2019, 10(1): 464. |
| [30] | Zou CS, Li LT, Miki D, et al. The genome of broomcorn millet [J]. Nat Commun, 2019, 10: 436. |
| [31] | Sun YL, Liu Y, Shi JF, et al. Biased mutations and gene losses underlying diploidization of the tetraploid broomcorn millet genome [J]. Plant J, 2023, 113(4): 787-801. |
| [32] | Lu Q, Zhao HN, Zhang ZQ, et al. Genomic variation in weedy and cultivated broomcorn millet accessions uncovers the genetic architecture of agronomic traits [J]. Nat Genet, 2024, 56(5): 1006-1017. |
| [1] | 罗稷林, 栗锦烨, 贾玉鑫. 马铃薯中重力响应调节基因鉴定及功能分析[J]. 生物技术通报, 2025, 41(6): 109-118. |
| [2] | 李旭娟, 李纯佳, 刘洪博, 徐超华, 林秀琴, 陆鑫, 刘新龙. 甘蔗腋芽形成发育过程的转录组分析[J]. 生物技术通报, 2025, 41(3): 202-218. |
| [3] | 岳丽昕, 王清华, 刘泽洲, 孔素萍, 高莉敏. 基于转录组和WGCNA筛选大葱雄性不育相关基因[J]. 生物技术通报, 2024, 40(9): 212-224. |
| [4] | 高萌萌, 赵天宇, 焦馨悦, 林春晶, 关哲允, 丁孝羊, 孙妍妍, 张春宝. 大豆细胞质雄性不育系及其恢复系的比较转录组分析[J]. 生物技术通报, 2024, 40(7): 137-149. |
| [5] | 白志元, 徐菲, 杨午, 王明贵, 杨玉花, 张海平, 张瑞军. 大豆细胞质雄性不育弱恢复型杂种F1育性转变的转录组分析[J]. 生物技术通报, 2024, 40(6): 134-142. |
| [6] | 秦健, 李振月, 何浪, 李俊玲, 张昊, 杜荣. 肌源性细胞分化的单细胞转录谱变化及细胞间通讯分析[J]. 生物技术通报, 2024, 40(6): 330-342. |
| [7] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [8] | 赵锐, 狄靖宜, 张广通, 刘浩, 高伟霞. 基于转录组学挖掘兽疫链球菌内源表达元件及高产透明质酸应用[J]. 生物技术通报, 2024, 40(10): 296-304. |
| [9] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [10] | 肖小军, 陈明, 韩德鹏, 余跑兰, 郑伟, 肖国滨, 周庆红, 周会汶. 甘蓝型油菜每角果粒数全基因组关联分析[J]. 生物技术通报, 2023, 39(3): 143-151. |
| [11] | 熊和丽, 沙茜, 刘韶娜, 相德才, 张斌, 赵智勇. 单细胞转录组测序技术在动物上的应用研究[J]. 生物技术通报, 2022, 38(3): 226-233. |
| [12] | 寇佳怡, 王玉玲, 曾睿琳, 兰道亮. 单细胞转录组测序技术及在哺乳动物上的应用[J]. 生物技术通报, 2022, 38(11): 41-48. |
| [13] | 郑青波, 叶娜, 张哓兰, 包鹏甲, 王福彬, 任稳稳, 廖月姣, 阎萍, 潘和平. 天祝白牦牛退行期毛囊细胞亚群鉴定以及特征基因生物信息学分析[J]. 生物技术通报, 2022, 38(10): 262-272. |
| [14] | 洪军, 卫夏怡, 吉冰洁, 叶延欣, 程天赐. 铜绿假单胞菌对鲎素耐药前后的差异表达基因及SNP变化研究[J]. 生物技术通报, 2021, 37(9): 191-202. |
| [15] | 陈建军, 赵怡迪, 曹香林. 脂多糖对鲤肠上皮细胞转录组模式的调控分析[J]. 生物技术通报, 2021, 37(8): 213-220. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||