








生物技术通报 ›› 2025, Vol. 41 ›› Issue (3): 202-218.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0755
• 研究报告 • 上一篇
李旭娟( ), 李纯佳, 刘洪博, 徐超华, 林秀琴, 陆鑫, 刘新龙(
), 李纯佳, 刘洪博, 徐超华, 林秀琴, 陆鑫, 刘新龙( )
)
收稿日期:2024-08-06
出版日期:2025-03-26
发布日期:2025-03-20
通讯作者:
刘新龙,男,博士,研究员,研究方向 :甘蔗分子育种;E-mail: lxlgood868@163.com作者简介:李旭娟,女,硕士,副研究员,研究方向 :甘蔗分子育种;E-mail: lixujuan2011@163.com
基金资助:
LI Xu-juan( ), LI Chun-jia, LIU Hong-bo, XU Chao-hua, LIN Xiu-qin, LU Xin, LIU Xin-long(
), LI Chun-jia, LIU Hong-bo, XU Chao-hua, LIN Xiu-qin, LU Xin, LIU Xin-long( )
)
Received:2024-08-06
Published:2025-03-26
Online:2025-03-20
摘要:
目的 发掘甘蔗腋芽形成发育关键代谢通路和调控基因,为甘蔗优良品种选育及遗传改良策略制定提供基因资源。 方法 以甘蔗品种‘新台糖22号(XTT22)’为材料,分别选取其幼嫩、半大、成型、成熟腋芽,以茎尖生长点为对照。首先通过RNA-seq获得甘蔗腋芽形成发育4个时期的转录组文库,然后使用DESeq2软件筛选差异表达基因(DEGs),并开展GO和KEGG功能富集分析,再通过RT-qPCR验证转录组测序结果的可靠性,最后通过WGCNA挖掘与甘蔗腋芽形成发育相关的核心基因。 结果 甘蔗腋芽形成发育4个时期共获得62 630个差异表达基因,KEGG分析主要富集在甘露糖型O-聚糖的生物合成、吲哚生物碱生物合成、油菜素内酯生物合成等通路;KEGG富集通路网络分析表明:植物激素信号转导、氰胺酸代谢以及苯丙烷类物质生物合成等通路是腋芽形成发育中共同富集的核心通路,而β-丙氨酸代谢和氮代谢是幼嫩腋芽发育期的特有核心通路;C-5支链二元酸代谢,色氨酸代谢和脂肪酸延生通路分别是成型芽和成熟芽特有富集核心通路。9个表达趋势变化明显的DEGs的RT-qPCR结果与RNA-seq 结果呈现相似的表达模式。WGCNA分析鉴定了UBR7、IRK和POLD等20个与甘蔗腋芽形成发育相关的核心基因。 结论 甘蔗腋芽形成发育过程中需要以甘露糖型O-聚糖、蔗糖、淀粉和氮素等作为物质和能量基础,同时还需要合成植物激素和次生代谢物来调控相关生长发育过程;UBR7、IRK以及POLD等基因可能在甘蔗腋芽形成发育中发挥正调控作用,而DGK1、PCaP1、LSD1等基因可能负调控甘蔗腋芽形成发育。
李旭娟, 李纯佳, 刘洪博, 徐超华, 林秀琴, 陆鑫, 刘新龙. 甘蔗腋芽形成发育过程的转录组分析[J]. 生物技术通报, 2025, 41(3): 202-218.
LI Xu-juan, LI Chun-jia, LIU Hong-bo, XU Chao-hua, LIN Xiu-qin, LU Xin, LIU Xin-long. Transcriptome Analysis of Axillary Bud Formation and Development in Sugarcane[J]. Biotechnology Bulletin, 2025, 41(3): 202-218.

图1 甘蔗茎尖生长点和不同发育阶段腋芽Bud0:茎尖生长点, Bud1:幼嫩腋芽, Bud2:半大发育腋芽, Bud3:成型发育腋芽, Bud4:成熟腋芽
Fig. 1 Stem apex and axillary buds of sugarcane at different developmental stagesBud0: Stem apex. Bud1: Tender axillary bud. Bud2: Semi-large axillary bud. Bud3: Young axillary bud. Bud4: Mature axillary bud
基因ID Gene ID | 基因名 Gene name | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|---|
| CL3565.Contig2_All | ARF7 | TTCGGTTGGAACATTGCTGC | TGTACAGGAGGGCAGACTGA |
| CL1829.Contig12_All | HSP70 | AGCTTGAGGGGATCTGCAAC | TGCAACACCACGTCTAACCA |
| Unigene43477_All | HSF-C2a | CTGTAGCACCGGAGACGAAG | CAGTTGGTGATGAGGGGCAA |
| CL6559.Contig7_All | PRP | CGTCCTCGACATGTGCTTCT | GTGATCATCAACCACGCACG |
| Unigene49447_All | SCL9 | TGGTCAGTCTTCTCAACCGC | GGGATCTGGCCATTCGTGAT |
| CL526.Contig2_All | WRKY48 | GCACTACCAGGAAGGCAGAG | CCTGCAGGTCCATTGGAGAG |
| CL11262.Contig6_All | MYB61 | GCATGTAGCTCGACCTTTTTGT | CCCAAGAGCTACGGACAGAG |
| Unigene48620_All | ATLP3 | TAGCACCGTCAACTTCGTCC | TGCACGCAGGCGTATAATCT |
| CL14664.Contig8_All | SAPK4 | ACCGTTTGTTTGCACTCTGC | TGGAAGAAATGCACACCCGA |
| GAPDH | CACGGCCACTGGAAGCA | TCCTCAGGGTTCCTGATGCC |
表1 实时荧光定量PCR引物
Table 1 Primers used for RT-qPCR
基因ID Gene ID | 基因名 Gene name | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|---|
| CL3565.Contig2_All | ARF7 | TTCGGTTGGAACATTGCTGC | TGTACAGGAGGGCAGACTGA |
| CL1829.Contig12_All | HSP70 | AGCTTGAGGGGATCTGCAAC | TGCAACACCACGTCTAACCA |
| Unigene43477_All | HSF-C2a | CTGTAGCACCGGAGACGAAG | CAGTTGGTGATGAGGGGCAA |
| CL6559.Contig7_All | PRP | CGTCCTCGACATGTGCTTCT | GTGATCATCAACCACGCACG |
| Unigene49447_All | SCL9 | TGGTCAGTCTTCTCAACCGC | GGGATCTGGCCATTCGTGAT |
| CL526.Contig2_All | WRKY48 | GCACTACCAGGAAGGCAGAG | CCTGCAGGTCCATTGGAGAG |
| CL11262.Contig6_All | MYB61 | GCATGTAGCTCGACCTTTTTGT | CCCAAGAGCTACGGACAGAG |
| Unigene48620_All | ATLP3 | TAGCACCGTCAACTTCGTCC | TGCACGCAGGCGTATAATCT |
| CL14664.Contig8_All | SAPK4 | ACCGTTTGTTTGCACTCTGC | TGGAAGAAATGCACACCCGA |
| GAPDH | CACGGCCACTGGAAGCA | TCCTCAGGGTTCCTGATGCC |
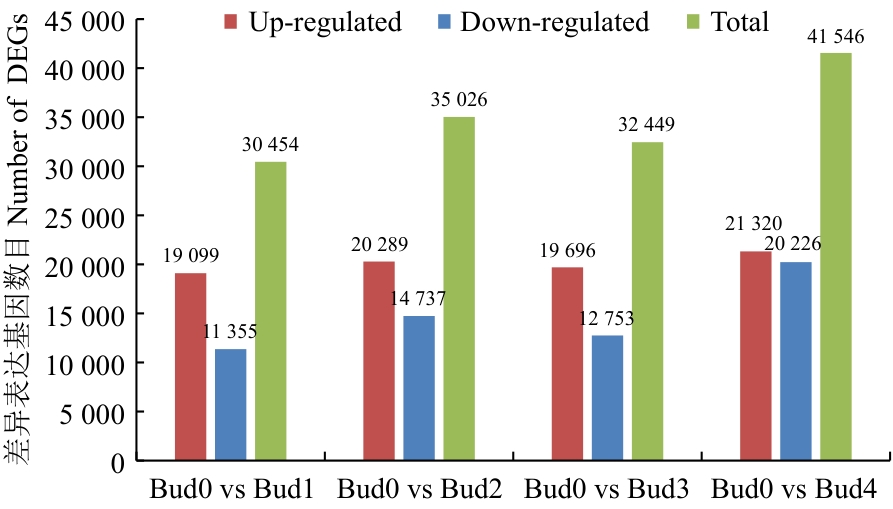
图3 甘蔗腋芽形成发育不同阶段差异表达基因数目统计
Fig. 3 Statistics of up-down-regulation genes between the sample groups at different stages of sugarcane axillary bud formation and development
| 基因编号 Gene ID | 基因名 Gene name | 基因注释 Gene annotation |
|---|---|---|
| CL3565.Contig2_All | ARF7 | XM_002453947.2:高粱生长素响应因子7,mRNA |
| CL1829.Contig12_All | HSP70 | XP_002456611.1:高粱热激蛋白70 mRNA |
| Unigene43477_All | HSF-C2a | XM_002451780.2:高粱热胁迫转录因子C-2a,转录变体X2,mRNA |
| CL6559.Contig7_All | PRP | CAB65536.1:玉米富含脯氨酸蛋白,mRNA |
| Unigene49447_All | SCL9 | XM_021446388.1:高粱SCL9蛋白(LOC110429790),转录变体X1,mRNA |
| Unigene48620_All | ATLP3 | XM_002450322.2:高粱生长素转运类蛋白3,mRNA |
| CL526.Contig2_All | WRKY48 | XM_002463811.2:高粱转录因子WRKY48 (LOC8086215),mRNA |
| CL11262.Contig6_All | MYB61 | XM_002457641.2:高粱转录因子MYB61 (LOC8078133),转录变体X1,mRNA |
| CL14664.Contig8_All | SAPK4 | XM_021447946.1:高粱丝氨酸/苏氨酸蛋白激酶SAPK4(LOC8058510),转录变体X3,mRNA |
表2 实时荧光定量 PCR验证候选基因
Table 2 Candidate genes for RT-qPCR
| 基因编号 Gene ID | 基因名 Gene name | 基因注释 Gene annotation |
|---|---|---|
| CL3565.Contig2_All | ARF7 | XM_002453947.2:高粱生长素响应因子7,mRNA |
| CL1829.Contig12_All | HSP70 | XP_002456611.1:高粱热激蛋白70 mRNA |
| Unigene43477_All | HSF-C2a | XM_002451780.2:高粱热胁迫转录因子C-2a,转录变体X2,mRNA |
| CL6559.Contig7_All | PRP | CAB65536.1:玉米富含脯氨酸蛋白,mRNA |
| Unigene49447_All | SCL9 | XM_021446388.1:高粱SCL9蛋白(LOC110429790),转录变体X1,mRNA |
| Unigene48620_All | ATLP3 | XM_002450322.2:高粱生长素转运类蛋白3,mRNA |
| CL526.Contig2_All | WRKY48 | XM_002463811.2:高粱转录因子WRKY48 (LOC8086215),mRNA |
| CL11262.Contig6_All | MYB61 | XM_002457641.2:高粱转录因子MYB61 (LOC8078133),转录变体X1,mRNA |
| CL14664.Contig8_All | SAPK4 | XM_021447946.1:高粱丝氨酸/苏氨酸蛋白激酶SAPK4(LOC8058510),转录变体X3,mRNA |

图5 腋芽形成发育4个阶段差异基因KEGG富集代谢通路气泡图A:幼嫩芽阶段;B:半大芽阶段;C:成型芽阶段;D:成熟芽阶段
Fig. 5 Bubble map of differential gene KEGG enrichment metabolic pathway in four stages of axillary bud formation and developmentA: The stage of tender axillary bud development; B: semi-large bud development stage; C: young bud development stage; D: mature bud development stage
模块 Module | 基因号 Gene ID | Nt功能注释 Nt Annotation |
|---|---|---|
| 蓝色 | CL11755.Contig1_All | XM_002437021.2:高粱E3泛素蛋白连接酶7 (LOC8072631),mRNA |
| CL11416.Contig2_All | XM_002467860.2:高粱LRR受体类丝氨酸/苏氨酸蛋白激酶IRK (LOC8082337),mRNA | |
| CL9946.Contig1_All | XM_002459710.2:高粱聚[adp-核糖]聚合酶1 (LOC8057336),mRNA | |
| Unigene27412_All | XM_002451536.2:高粱DNA聚合酶δ催化亚基POLD (LOC8074668),mRNA | |
| CL414.Contig1_All | XM_002437978.2:高粱LRR受体样丝氨酸/苏氨酸蛋白激酶ERECTA (LOC8077321),mRNA | |
| 褐色 | CL9713.Contig1_All | M86717.1:甘蔗核酮糖1,5-二磷酸羧化酶/加氧酶小亚基基因 |
| CL7506.Contig2_All | XM_021463606.1:高粱豌豆蛋白(LOC8064511),转录变体X2,mRNA | |
| CL1675.Contig6_All | XM_021461443.1:高粱LNK1-类蛋白(LOC110435648),mRNA | |
| CL10608.Contig2_All | NM_001153244.1:玉米热胁迫转录因子A-6b (cl30385_1),mRNA | |
| Unigene37307_All | XM_002437010.2:高粱转录因子WRKY19,mRNA | |
| 绿色 | CL11922.Contig2_All | 芒草ARM重复超家族蛋白(NCGR_LOCUS6946),mRNA |
| CL4984.Contig2_All | XM_002456186.2:高粱单myb4组蛋白 (LOC8069770),mRNA | |
| Unigene34707_All | XM_008679969.2:玉米VIN3-类蛋白 (LOC103652991),mRNA | |
| Unigene7438_All | XM_002439022.2:高粱胼胝质合成酶3 (LOC8066386),mRNA | |
| CL6695.Contig6_All | XM_002462606.2:高粱adp核糖化因子gtpase激活蛋白AGD3 (LOC8054803),mRNA | |
| 黑色 | CL471.Contig5_All | XM_002443369.2:高粱二酰基甘油激酶 (LOC8074256),mRNA |
| CL654.Contig6_All | KT891986.1:甘蔗质膜阳离子结合蛋白(pcap1),mRNA | |
| CL5540.Contig5_All | XM_002445052.2:高粱LSD1蛋白,XM_002445052.2,mRNA | |
| CL5050.Contig15_All | XM_002454463.2:高粱水通道蛋白PIP1-2,mRNA | |
| Unigene46867_All | XM_021450410.1:高粱δ(24)-类甾醇还原酶(LOC110431358),mRNA |
表3 WGCNA候选基因及其注释
Table 3 WGCNA candidate genes and their annotations
模块 Module | 基因号 Gene ID | Nt功能注释 Nt Annotation |
|---|---|---|
| 蓝色 | CL11755.Contig1_All | XM_002437021.2:高粱E3泛素蛋白连接酶7 (LOC8072631),mRNA |
| CL11416.Contig2_All | XM_002467860.2:高粱LRR受体类丝氨酸/苏氨酸蛋白激酶IRK (LOC8082337),mRNA | |
| CL9946.Contig1_All | XM_002459710.2:高粱聚[adp-核糖]聚合酶1 (LOC8057336),mRNA | |
| Unigene27412_All | XM_002451536.2:高粱DNA聚合酶δ催化亚基POLD (LOC8074668),mRNA | |
| CL414.Contig1_All | XM_002437978.2:高粱LRR受体样丝氨酸/苏氨酸蛋白激酶ERECTA (LOC8077321),mRNA | |
| 褐色 | CL9713.Contig1_All | M86717.1:甘蔗核酮糖1,5-二磷酸羧化酶/加氧酶小亚基基因 |
| CL7506.Contig2_All | XM_021463606.1:高粱豌豆蛋白(LOC8064511),转录变体X2,mRNA | |
| CL1675.Contig6_All | XM_021461443.1:高粱LNK1-类蛋白(LOC110435648),mRNA | |
| CL10608.Contig2_All | NM_001153244.1:玉米热胁迫转录因子A-6b (cl30385_1),mRNA | |
| Unigene37307_All | XM_002437010.2:高粱转录因子WRKY19,mRNA | |
| 绿色 | CL11922.Contig2_All | 芒草ARM重复超家族蛋白(NCGR_LOCUS6946),mRNA |
| CL4984.Contig2_All | XM_002456186.2:高粱单myb4组蛋白 (LOC8069770),mRNA | |
| Unigene34707_All | XM_008679969.2:玉米VIN3-类蛋白 (LOC103652991),mRNA | |
| Unigene7438_All | XM_002439022.2:高粱胼胝质合成酶3 (LOC8066386),mRNA | |
| CL6695.Contig6_All | XM_002462606.2:高粱adp核糖化因子gtpase激活蛋白AGD3 (LOC8054803),mRNA | |
| 黑色 | CL471.Contig5_All | XM_002443369.2:高粱二酰基甘油激酶 (LOC8074256),mRNA |
| CL654.Contig6_All | KT891986.1:甘蔗质膜阳离子结合蛋白(pcap1),mRNA | |
| CL5540.Contig5_All | XM_002445052.2:高粱LSD1蛋白,XM_002445052.2,mRNA | |
| CL5050.Contig15_All | XM_002454463.2:高粱水通道蛋白PIP1-2,mRNA | |
| Unigene46867_All | XM_021450410.1:高粱δ(24)-类甾醇还原酶(LOC110431358),mRNA |
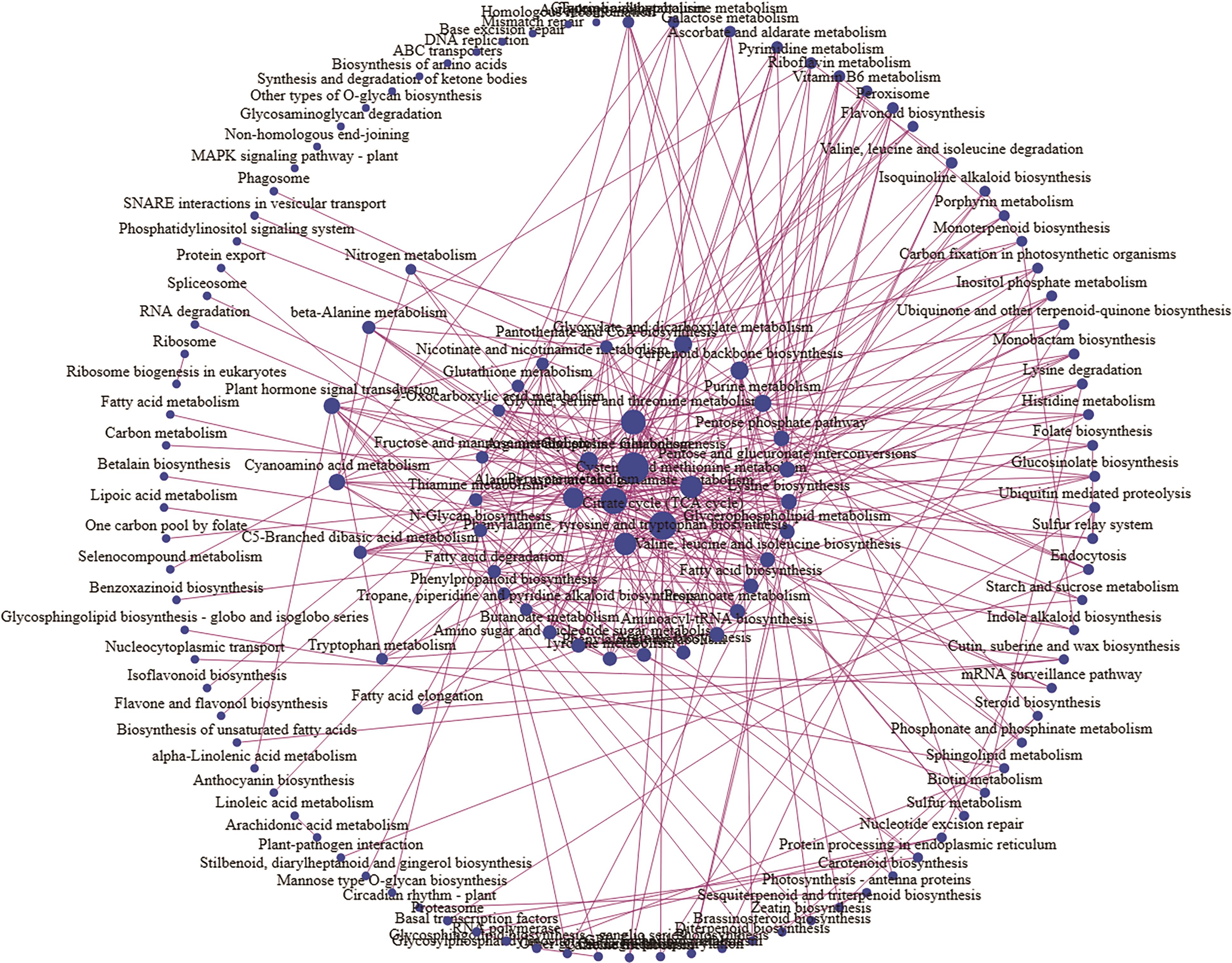
图6 甘蔗腋芽形成发育差异基因涉及的132个KEGG通路网络关系图红色箭头为共有和特有富集通路中的核心通路
Fig. 6 Network diagram of 132 KEGG pathways involved in differential genes of axillary bud formation and development in sugarcaneThe red arrows are the core pathways in the common and specific enrichment pathways

图7 基因共表达网络构建结果A:软阈值确定;B:基因聚类和模块构建;C:样本与模块相关性热图(红色格子代表样本与模块具有正相关性,紫色格子代表样本与模块具有负相关性,图中括号内外的数值分别为 P 值和相关系数r)
Fig. 7 Result of gene co-expression network constructionA: The determination of soft threshold. B: Gene clustering and module construction. C: Heat map of the correlation between modules and samples (Red color of each box represents the positive correlation between samples and module. Purple color of each box represents the negative relationships between samples and module. The values inside and outside the brackets in the figure are P-values and correlation coefficients, respectively)

图8 关键模块内核心基因的互作网络图节点和标签大小代表基因的连通度。A:蓝色模块;B:棕色模块;C:绿色模块;D:黑色模块
Fig. 8 Gene networks of hub genes of key modulesThe node and label size indicates the gene connectivity. A: Blue module. B: Brown module. C: Green module. D: Black module
| 1 | Islam MS, Yang XP, Sood S, et al. Molecular characterization of genetic basis of Sugarcane Yellow Leaf Virus (SCYLV) resistance in Saccharum spp. hybrid [J]. Plant Breed, 2018, 137(4): 598-604. |
| 2 | Oz MT, Altpeter A, Karan R, et al. CRISPR/Cas9-mediated multi-allelic gene targeting in sugarcane confers herbicide tolerance [J]. Front Genome Ed, 2021, 3: 673566. |
| 3 | 陆鑫, 毛钧, 刘新龙, 等. 甘蔗双腋芽突变体表型鉴定及应用潜力评估 [J]. 植物遗传资源学报, 2020, 21(2): 314-320. |
| Lu X, Mao J, Liu XL, et al. Phenotypic identification and evaluation of application potential in double axillary buds mutant of sugarcane [J]. J Plant Genet Resour, 2020, 21(2): 314-320. | |
| 4 | 张跃彬. 现代甘蔗糖业 [M]. 北京: 科学出版社, 2013. |
| Zhang YB. Modern sugar industry [M]. Beijing: Science Press, 2013. | |
| 5 | Jürgens G. Apical-basal pattern formation in Arabidopsis embryogenesis [J]. EMBO J, 2001, 20(14): 3609-3616. |
| 6 | Long J, Barton MK. Initiation of axillary and floral meristems in Arabidopsis [J]. Dev Biol, 2000, 218(2): 341-353. |
| 7 | Tanaka W, Ohmori Y, Ushijima T, et al. Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1 [J]. Plant Cell, 2015, 27(4): 1173-1184. |
| 8 | Liu Y, Ding YF, Wang QS, et al. Effect of plant growth regulators on the growth of rice tiller bud and the changes of endogenous hormones [J]. Acta Agron Sin, 2011, 37(4): 670-676. |
| 9 | Ortiz-Morea FA, Vicentini R, Silva GFF, et al. Global analysis of the sugarcane microtranscriptome reveals a unique composition of small RNAs associated with axillary bud outgrowth [J]. J Exp Bot, 2013, 64(8): 2307-2320. |
| 10 | Yan HF, Zhou HW, Luo HM, et al. Characterization of full-length transcriptome in Saccharum officinarum and molecular insights into tiller development [J]. BMC Plant Biol, 2021, 21(1): 228. |
| 11 | Vasantha S, Shekinah DE, Gupta C, et al. Tiller production, regulation and senescence in sugarcane (Saccharum species hybrid) Genotypes [J]. Sugar Tech, 2012, 14(2): 156-160. |
| 12 | 叶燕萍. 乙烯利促进甘蔗有效分蘖的生理生化机理研究 [D]. 南宁: 广西大学, 2006. |
| Ye YP. Studies on the physiological and biochemical mechanism on ethephon promoting effective tillering in sugarcane [D]. Nanning: Guangxi University, 2006. | |
| 13 | 王威豪, 王一丁, 莫云川, 等. 水分胁迫下喷施乙烯利对甘蔗分蘖及农艺性状的影响 [J]. 广西农业科学, 2007, 38(2): 148-151. |
| Wang WH, Wang YD, Mo YC, et al. Effects of ethephon on tillering and agronomic characters in sugarcane under water-stress [J]. Guangxi Agric Sci, 2007, 38(2): 148-151. | |
| 14 | 吴凯朝. 乙烯利促进甘蔗分蘖生理与形态学效应的研究 [D]. 南宁: 广西大学, 2005. |
| Wu KC. Studies on physiological and morphological effects of ethephon on tillering of sugarcane [D]. Nanning: Guangxi University, 2005. | |
| 15 | 李旭娟, 李纯佳, 徐超华, 等. 甘蔗MOC1基因(ScMOC1)的克隆与表达分析 [J]. 植物遗传资源学报, 2017, 18(4): 734-746. |
| Li XJ, Li CJ, Xu CH, et al. Cloning and expression analysis of the MOC1 gene(ScMOC1) in sugarcane(Saccharum officinarum) [J]. J Plant Genet Resour, 2017, 18(4): 734-746. | |
| 16 | 李旭娟, 林秀琴, 刘洪博, 等. 甘蔗TB1基因的克隆与生物信息学分析 [J]. 热带作物学报, 2015, 36(11): 1978-1985. |
| Li XJ, Lin XQ, Liu HB, et al. Cloning and bioinformatics analysis of the TB1 gene in sugarcane [J]. Chin J Trop Crops, 2015, 36(11): 1978-1985. | |
| 17 | 李旭娟, 刘洪博, 林秀琴, 等. 甘蔗KNOX基因(Sckn1)的电子克隆及生物信息学分析 [J]. 基因组学与应用生物学, 2015, 34(1): 136-142. |
| Li XJ, Liu HB, Lin XQ, et al. In silico cloning and bioinformatics analysis of KNOX gene in sugarcane (Sckn1) [J]. Genom Appl Biol, 2015, 34(1): 136-142. | |
| 18 | Wang LK, Feng ZX, Wang X, et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data [J]. Bioinformatics, 2010, 26(1): 136-138. |
| 19 | Ling H, Wu QB, Guo JL, et al. Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR [J]. PLoS One, 2014, 9(5): e97469. |
| 20 | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC T method [J]. Methods, 2001, 25(4): 402-408. |
| 21 | Singh P, Song QQ, Singh RK, et al. Proteomic analysis of the resistance mechanisms in sugarcane during Sporisorium scitamineum infection [J]. Int J Mol Sci, 2019, 20(3): 569. |
| 22 | 钱甫, 张占琴, 陈树宾, 等. 基于GWAS和WGCNA分析挖掘玉米花期相关候选基因 [J]. 作物学报, 2023, 49(12): 3261-3276. |
| Qian F, Zhang ZQ, Chen SB, et al. Mining maize flowering traits related candidate genes based on GWAS and WGCNA data [J]. Acta Agron Sin, 2023, 49(12): 3261-3276. | |
| 23 | Leyser O. The control of shoot branching: an example of plant information processing [J]. Plant Cell Environ, 2009, 32(6): 694-703. |
| 24 | Chen D, Lu X, Wu XY, et al. Transcriptome analysis of axillary bud differentiation in a new dual-axillary bud genotype of sugarcane [J]. Genet Resour Crop Evol, 2020, 67(3): 685-701. |
| 25 | 吴佳, 李惠中, 邓权清, 等. 甘蔗鞭黑粉菌致病力差异菌株转录组分析 [J]. 华中农业大学学报, 2020, 39(3): 54-59. |
| Wu J, Li HZ, Deng QQ, et al. Analyzing transcriptome of Sporisorium scitamineum strains with different pathogenicity [J]. J Huazhong Agric Univ, 2020, 39(3): 54-59. | |
| 26 | 许跃语. 甘蔗响应黄叶病毒侵染的转录组差异表达研究 [D]. 福州: 福建农林大学, 2019. |
| Xu YY. Transcriptomic analysis of differentially expressed genes in sugarcane in response to yellow leaf virus infection [D]. Fuzhou: Fujian Agriculture and Forestry University, 2019. | |
| 27 | 苏亚春. 甘蔗应答黑穗病菌侵染的转录组与蛋白组研究及抗性相关基因挖掘 [D]. 福州: 福建农林大学, 2014. |
| Su YC. Study on transcriptome and proteome of sugarcane in response to Ustilago scitamella infection and gene mining related to resistance [D]. Fuzhou: Fujian Agriculture and Forestry University, 2014. | |
| 28 | 丘立杭, 罗含敏, 陈荣发, 等. 基于RNA-Seq的甘蔗主茎和分蘖茎转录组建立及初步分析 [J]. 基因组学与应用生物学, 2018, 37(3): 1271-1279. |
| Qiu LH, Luo HM, Chen RF, et al. Establishment and preliminary analysis on transcriptome of sugarcane between stalk and tiller based on RNA-seq technology [J]. Genom Appl Biol, 2018, 37(3): 1271-1279. | |
| 29 | Dapanage M, Bhat S. Transcriptome profiling through next-generation sequencing in sugarcane under moisture-deficit stress condition [J]. Sugar Tech, 2020, 22(3): 396-410. |
| 30 | 刘洪博, 刘新龙, 苏火生, 等. 干旱胁迫下割手密根系转录组差异表达分析 [J]. 中国农业科学, 2017, 50(6): 1167-1178. |
| Liu HB, Liu XL, Su HS, et al. Transcriptome difference analysis of Saccharum spontaneum roots in response to drought stress [J]. Sci Agric Sin, 2017, 50(6): 1167-1178. | |
| 31 | 张珂. 甘蔗腋芽萌发相关植物激素代谢分析及促萌发诱导剂的筛选 [D]. 福州: 福建农林大学, 2020. |
| Zhang K. Metabolic analysis of plant hormones related to germination of sugarcane axillary buds and screening of germination inducer [D]. Fuzhou: Fujian Agriculture and Forestry University, 2020. | |
| 32 | Jin JP, Tian F, Yang DC, et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants [J]. Nucleic Acids Res, 2017, 45(D1): 1040-1045. |
| 33 | Xiong SF, Zhao YX, Chen YC, et al. Transcriptome and proteome analyses reveal a critical role of the sucrose anabolic pathway in regulating Quercus fabri Hance axillary bud outgrowth [J]. Ind Crops Prod, 2023, 194: 116284. |
| 34 | 胡鑫, 罗正英, 李纯佳, 等. 基于二代和三代转录组测序揭示甘蔗重要亲本对黑穗病菌侵染的响应机制 [J]. 作物学报, 2023, 49(9): 2412-2432. |
| Hu X, Luo ZY, Li CJ, et al. Comparative transcriptome analysis of elite ‘ROC’ sugarcane parents for exploring genes involved in Sporisorium scitamineum infection by using Illumina-and SMRT-based RNA-seq [J]. Acta Agron Sin, 2023, 49(9): 2412-2432. | |
| 35 | 韦婉羚, 杨海霞, 何文, 等. 基于转录组测序的赤苍藤根、茎和叶基因表达分析 [J]. 植物生理学报, 2023, 59(2): 333-344. |
| Wei WL, Yang HX, He W, et al. Transcriptiomic data analysis of roots, stems, and leaves of Erythropalum scandens [J]. Plant Physiol J, 2023, 59(2): 333-344. | |
| 36 | 张海青, 张恒涛, 高启明, 等. 转录组分析筛选调控苹果分枝能力的关键基因 [J]. 中国农业科学, 2024, 57(10): 1995-2009. |
| Zhang HQ, Zhang HT, Gao QM, et al. Transcriptome analysis for screening key genes related to regulating branching ability in apple [J]. Sci Agric Sin, 2024, 57(10): 1995-2009. | |
| 37 | 孙晓琛, 原静静, 栗锦鹏, 等. 干旱胁迫对党参苯丙素及倍半萜和三萜类成分合成相关基因的调控 [J]. 中成药, 2022, 44(12): 3902-3909. |
| Sun XC, Yuan JJ, Li JP, et al. Genetic adaptation under drought stress for the synthesis of phenylpropanoid, sesquiterpenoids and triterpenoids in Codonopsis pilosula [J]. Chin Tradit Pat Med, 2022, 44(12): 3902-3909. | |
| 38 | 郝瑞杰, 邱晨, 耿晓云, 等. 梅花Pm ABCG9在苯甲醇挥发中的功能分析 [J]. 中国农业科学, 2023, 56(13): 2574-2585. |
| Hao RJ, Qiu C, Geng XY, et al. The function of Pm ABCG9 transporter related to the volatilization of benzyl alcohol in Prunus mume [J]. Sci Agric Sin, 2023, 56(13): 2574-2585. | |
| 39 | 荐红举, 张梅花, 尚丽娜, 等. 利用WGCNA筛选马铃薯块茎发育候选基因 [J]. 作物学报, 2022, 48(7): 1658-1668. |
| Jian HJ, Zhang MH, Shang LN, et al. Screening candidate genes involved in potato Tuber development using WGCNA [J]. Acta Agron Sin, 2022, 48(7): 1658-1668. | |
| 40 | 张恒燕, 张木清. 基于WGCNA挖掘外源GA3调节甘蔗节间伸长相关基因 [J]. 分子植物育种,2022:1-21. |
| Zhang HY, Zhang MQ. Mining of genes related to exogenous GA3 regulation of sugarcane internode elongation based on WGCNA [J]. Molecular Plant Breeding, 2022:1-21. | |
| 41 | 李佩婷, 赵振丽, 黄潮华, 等. 基于转录组及WGCNA的甘蔗干旱响应调控网络分析 [J]. 作物学报, 2022, 48(7): 1583-1611. |
| Li PT, Zhao ZL, Huang CH, et al. Analysis of drought responsive regulatory network in sugarcane based on transcriptome and WGCNA [J]. Acta Agron Sin, 2022, 48(7): 1583-1611. | |
| 42 | Barbier FF, Lunn JE, Beveridge CA. Ready, steady, go! A sugar hit starts the race to shoot branching [J]. Curr Opin Plant Biol, 2015, 25: 39-45. |
| 43 | Barbier FF, Dun EA, Kerr SC, et al. An update on the signals controlling shoot branching [J]. Trends Plant Sci, 2019, 24(3): 220-236. |
| 44 | Barbier F, Péron T, Lecerf M, et al. Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida [J]. J Exp Bot, 2015, 66(9): 2569-2582. |
| 45 | Zheng YY, Zhang SS, Luo YQ, et al. Rice OsUBR7 modulates plant height by regulating histone H2B monoubiquitination and cell proliferation [J]. Plant Commun, 2022, 3(6): 100412. |
| 46 | 张晓晶. 拟南芥二酰基甘油激酶AtDGKs在生长发育中的功能分析 [D]. 广州: 华南农业大学, 2017. |
| Zhang XJ. Functional analysis of a DGK gene encoding a diacylglycerol kinase in Arabidopsis [D]. Guangzhou: South China Agricultural University, 2017. | |
| 47 | 童君, 陈景光, 朱静雯, 等. 水稻 C2C2锌指蛋白基因OsLSD2对日本晴品种生长及氮素利用的影响 [J]. 中国水稻科学, 2014, 28(4): 435-441. |
| Tong J, Chen JG, Zhu JW, et al. Effects of rice C2C2 zinc finger protein gene OsLSD2 on the growth and nitrogen utilization of nipponbare [J]. Chin J Rice Sci, 2014, 28(4): 435-441. |
| [1] | 岳丽昕, 王清华, 刘泽洲, 孔素萍, 高莉敏. 基于转录组和WGCNA筛选大葱雄性不育相关基因[J]. 生物技术通报, 2024, 40(9): 212-224. |
| [2] | 高萌萌, 赵天宇, 焦馨悦, 林春晶, 关哲允, 丁孝羊, 孙妍妍, 张春宝. 大豆细胞质雄性不育系及其恢复系的比较转录组分析[J]. 生物技术通报, 2024, 40(7): 137-149. |
| [3] | 王超敏, 何美丹, 王文治, 袁潜华, 张树珍, 沈林波. 甘蔗条点病毒荧光定量PCR检测方法的建立及应用[J]. 生物技术通报, 2024, 40(6): 126-133. |
| [4] | 白志元, 徐菲, 杨午, 王明贵, 杨玉花, 张海平, 张瑞军. 大豆细胞质雄性不育弱恢复型杂种F1育性转变的转录组分析[J]. 生物技术通报, 2024, 40(6): 134-142. |
| [5] | 秦健, 李振月, 何浪, 李俊玲, 张昊, 杜荣. 肌源性细胞分化的单细胞转录谱变化及细胞间通讯分析[J]. 生物技术通报, 2024, 40(6): 330-342. |
| [6] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [7] | 田春艳, 李旭娟, 李纯佳, 毛钧, 刘新龙. 甘蔗属种及其近缘属种蔗茅的全基因组密码子偏好性分析[J]. 生物技术通报, 2024, 40(3): 202-214. |
| [8] | 辛奇, 李压凡, 尹铮, 张晓丹, 陈霆, 刘晓华. 甘蔗CBL-CIPK基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(2): 197-211. |
| [9] | 赵锐, 狄靖宜, 张广通, 刘浩, 高伟霞. 基于转录组学挖掘兽疫链球菌内源表达元件及高产透明质酸应用[J]. 生物技术通报, 2024, 40(10): 296-304. |
| [10] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [11] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [12] | 李心怡, 姜春秀, 薛丽, 蒋洪涛, 姚伟, 邓祖湖, 张木清, 余凡. 多荧光标记引物增强甘蔗染色体寡聚核苷酸探针杂交信号[J]. 生物技术通报, 2023, 39(5): 103-111. |
| [13] | 罗艳菊, 谢林艳, 邹清林, 李四杰, 刘涵, 刘鲁峰, 何丽莲, 李富生. 内生菌对干旱胁迫下甘蔗的生理响应及抗旱性评价[J]. 生物技术通报, 2023, 39(12): 219-228. |
| [14] | 黄佳艳, 冯小艳, 沈林波, 王文治, 胡海燕, 张树珍. 甘蔗ShPR10基因的克隆及其编码蛋白与甘蔗线条花叶病毒P1蛋白的互作研究[J]. 生物技术通报, 2023, 39(10): 163-174. |
| [15] | 徐扬, 丁红, 张冠初, 郭庆, 张智猛, 戴良香. 盐胁迫下花生种子萌发期代谢组学分析[J]. 生物技术通报, 2023, 39(1): 199-213. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||