








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 1-12.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0953
• 综述与专论 • 下一篇
淦晨露1( ), 游雨婷1, 谢菡萏1,2, 曾子贤1,2, 朱博1,2(
), 游雨婷1, 谢菡萏1,2, 曾子贤1,2, 朱博1,2( )
)
收稿日期:2025-09-05
出版日期:2026-01-26
发布日期:2026-02-04
通讯作者:
朱博,女,教授,研究方向 :植物功能基因组学、植物抗逆转录调控;E-mail: bozhu@sicnu.edu.cn作者简介:淦晨露,女,硕士研究生,研究方向 :植物非生物胁迫;E-mail: gancl34@163.com
基金资助:
GAN Chen-lu1( ), YOU Yu-ting1, XIE Han-dan1,2, ZENG Zi-xian1,2, ZHU Bo1,2(
), YOU Yu-ting1, XIE Han-dan1,2, ZENG Zi-xian1,2, ZHU Bo1,2( )
)
Received:2025-09-05
Published:2026-01-26
Online:2026-02-04
摘要:
植物黄素单加氧酶(flavin monooxygenases, FMOs)是一类以黄素腺嘌呤二核苷酸(FAD)为辅因子的氧化还原酶,能够催化多种底物的氧化反应,在植物代谢调控和环境适应中发挥重要作用。植物FMOs超家族包含多个功能亚家族,其中以FMO1、YUCCAs和FMOGS-OXs最为典型,分别在免疫防御、生长素合成、次生代谢及逆境响应等核心过程中扮演关键角色。本文系统梳理了这3个亚家族的结构特征、进化分化与功能机制,并总结了其在植物生长发育和逆境胁迫中的最新研究进展。研究表明,FMOs家族普遍含有保守的FAD/NADPH结合结构域,且在进化过程中通过基因扩增实现功能多样化,同时展现出显著的功能冗余与多效性特征。尽管如此,FMOs家族的天然底物谱尚未系统解析,其基因扩张与进化的分子机制有待深入阐明。同时,其时空特异性及功能冗余性严重限制了其分子功能和应用价值的深入研究。未来,随着代谢组学、空间组学和人工智能等前沿技术的发展,FMOs功能解析与应用转化将迎来新机遇,有望为作物抗逆、提产的分子设计育种提供新的靶点。
淦晨露, 游雨婷, 谢菡萏, 曾子贤, 朱博. 植物黄素单加氧酶研究进展[J]. 生物技术通报, 2026, 42(1): 1-12.
GAN Chen-lu, YOU Yu-ting, XIE Han-dan, ZENG Zi-xian, ZHU Bo. Research Progress in Flavin Monooxygenases in Plants[J]. Biotechnology Bulletin, 2026, 42(1): 1-12.
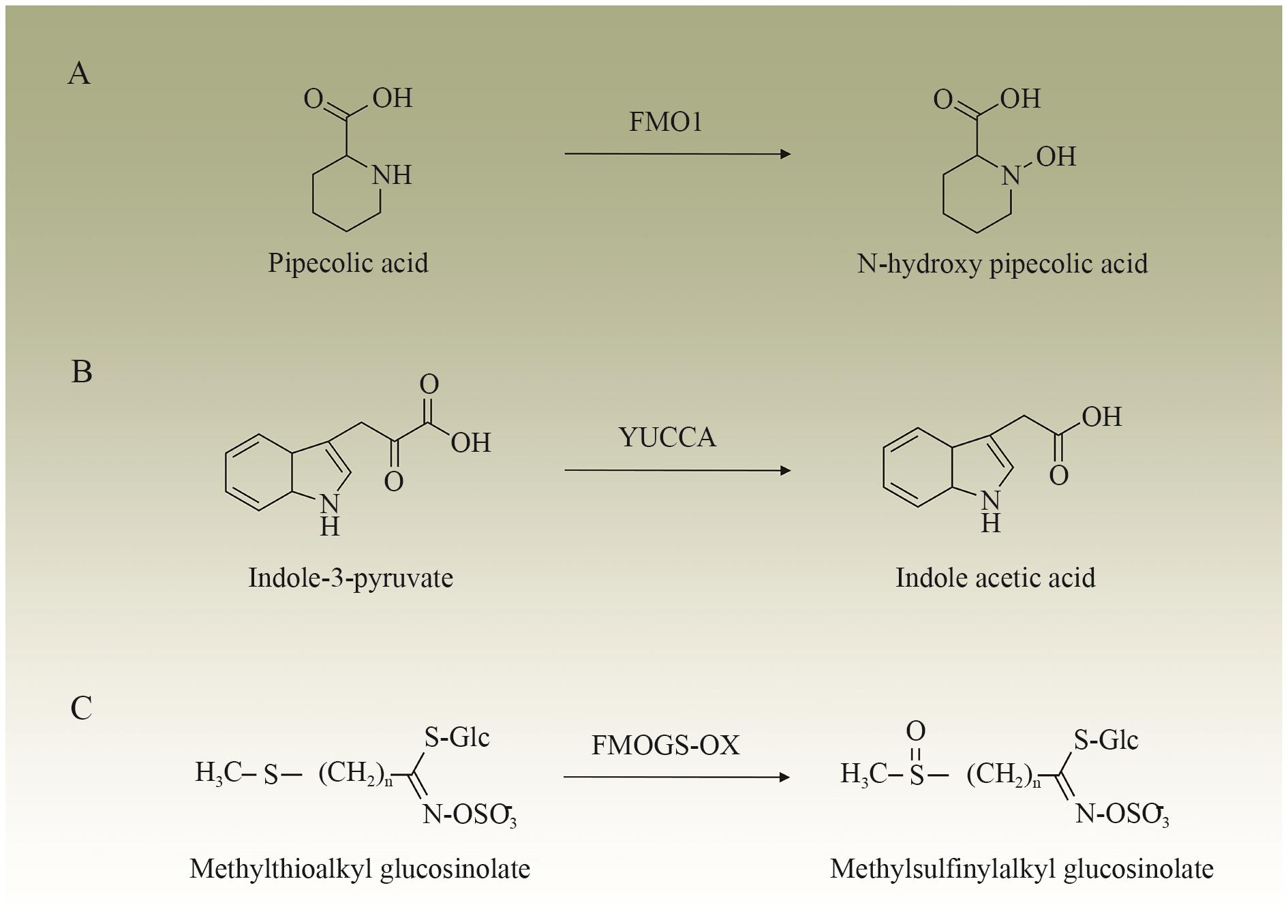
图2 植物黄素单加氧酶催化反应A: FMO1催化哌啶甲酸转变为N-羟基哌啶甲酸;B: YUCCAs催化吲哚-3-丙酮酸转变为吲哚乙酸;C: FMOGS-OXs催化甲硫基烷基硫代葡萄糖苷转变为甲基亚磺酰基烷基硫代葡萄糖苷
Fig. 2 Catalytic reaction of flavin monooxygenases in plantsA: FMO1 catalyzes the transformation of pipecolic acid to N-hydroxy piperidine pipecolic acid. B: YUCCAs catalyze the conversion of indole-3-pyruvate to indole acetic acid. C: FMOGS-OXs catalyze the conversion of methylthioalkyl glucosinolates to methylsulfinylalkyl glucosinolates
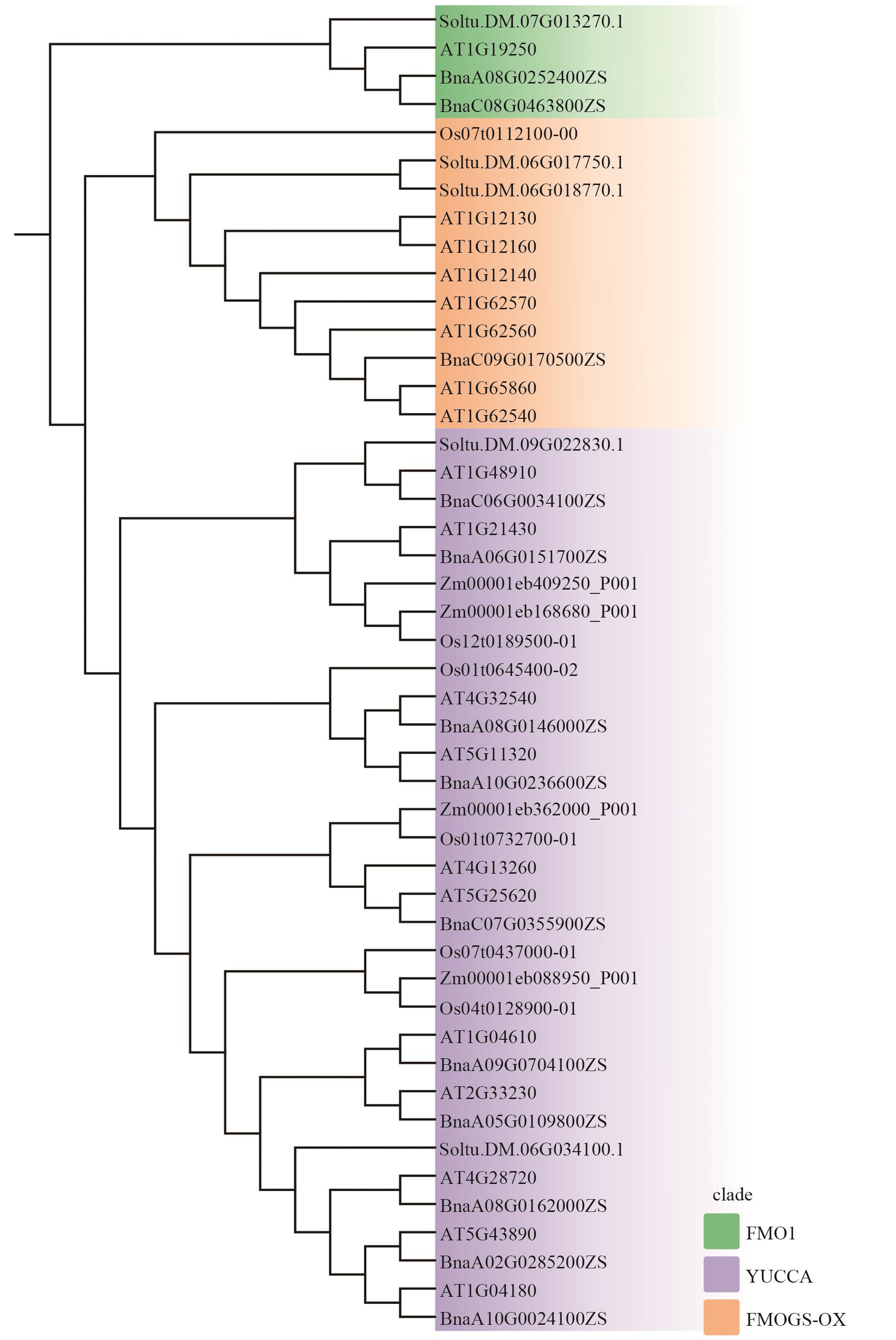
图3 拟南芥、油菜、水稻、玉米和马铃薯黄素单加氧酶FMOs系统发育树绿色:FMO1;紫色:YUCCAs;橙色:FMOGS-OXs;拟南芥(Arabidopsis thaliana)At;油菜(Brassica napus)Bna;水稻(Oryza sativa)Os;玉米(Zea mays)Zm;马铃薯(Solanum tuberosum)St
Fig. 3 Phyleigenic trees of flavin monooxygenases in Arabidopsis thaliana, Brassica napus, Oryza sativa, Zea mays, and Solanum tuberosumGreen: FMO1. Purple: YUCCAs. Orange: FMOGS-OXs. At (Arabidopsis thaliana); Bna (Brassica napus); Os (Oryza sativa); Zm (Zea mays); St (Solanum tuberosum)
生理过程 Physiological process | 蛋白类别 Protein category | 植物种类 Plant species | 基因名称 Gene name | 功能描述 Function | 参考文献 References |
|---|---|---|---|---|---|
生长发育 Growth and development | FMO1 | 拟南芥 | FMO1 | 根的发育 | [ |
| 其他 | TaFMO1-5B | 根的发育 | [ | ||
| YUCCAs | 拟南芥 | YUC2、YUC5、YUC8、YUC9 | 茎、叶、花序发育 | [ | |
| 水稻 | OsYUC1、OsYUC2、OsYUC11 | 冠根、籽粒、胚乳发育 | [ | ||
| 玉米 | spi1 | 腋生分生组织的形成 | [ | ||
| 其他 | SiYUC2、SiYUC6、SiYUC8、SiYUC11、Turnera YUC6、FvYUC6、EbYUC2、PpYUC11、CsYUC10、CmYUC6、CmYUC11 | 雄配子体、花粉、穗子、果实、叶片发育及胚胎形成 | [ | ||
| FMO1 | 拟南芥 | FMO1 | 光胁迫 | [ | |
| 其他 | FMO1 | 干旱胁迫 | [ | ||
非生物胁迫 Abiotic stress | YUCCAs | 拟南芥 | YUC6、YUC8 | 干旱、高温、重金属胁迫 | [ |
| 其他 | CsYUC8/9、CsYUC10b | 高温、低温、盐胁迫 | [ | ||
| FMOGS-OXs | 拟南芥 | FMOGS-OX2-7 | 干旱、低温、盐、渗透胁迫及激素处理 | [ | |
| 其他 | TsFMO | 盐胁迫 | [ | ||
生物胁迫 Biological stress | FMO1 | 拟南芥 | FMO1 | 系统性获得抗性(SAR)、降解病原毒素 | [ |
| 其他 | FMO | NHP合成、抑制黄酮合成并促进木质素积累 | [ | ||
| YUCCAs | 其他 | Bs3 | 触发超敏反应 | [ | |
| FMOGS-OXs | 拟南芥 | FMOGS-OXs | GSL生物合成 | [ | |
次生代谢 Secondary metabolism | FMOGS-OXs | 拟南芥 | FMOGS-OXs | GSL生物合成 | [ |
| 其他 | FMOGS-OX2、BrrFMOGS-OX2、BrrFMOGS-OX5.1、BrrFMOGS-OX5.2、BrrFMOGS-OX6.1、BrrFMOGS-OX6.2、FMOGS-OXs、AsFMO1 | GSL生物合成、催化蒜氨酸S-氧化 | [ |
表1 不同物种中3类黄素单加氧酶的生理功能
Table 1 Physiological functions of three types of flavin monooxygenases in different species
生理过程 Physiological process | 蛋白类别 Protein category | 植物种类 Plant species | 基因名称 Gene name | 功能描述 Function | 参考文献 References |
|---|---|---|---|---|---|
生长发育 Growth and development | FMO1 | 拟南芥 | FMO1 | 根的发育 | [ |
| 其他 | TaFMO1-5B | 根的发育 | [ | ||
| YUCCAs | 拟南芥 | YUC2、YUC5、YUC8、YUC9 | 茎、叶、花序发育 | [ | |
| 水稻 | OsYUC1、OsYUC2、OsYUC11 | 冠根、籽粒、胚乳发育 | [ | ||
| 玉米 | spi1 | 腋生分生组织的形成 | [ | ||
| 其他 | SiYUC2、SiYUC6、SiYUC8、SiYUC11、Turnera YUC6、FvYUC6、EbYUC2、PpYUC11、CsYUC10、CmYUC6、CmYUC11 | 雄配子体、花粉、穗子、果实、叶片发育及胚胎形成 | [ | ||
| FMO1 | 拟南芥 | FMO1 | 光胁迫 | [ | |
| 其他 | FMO1 | 干旱胁迫 | [ | ||
非生物胁迫 Abiotic stress | YUCCAs | 拟南芥 | YUC6、YUC8 | 干旱、高温、重金属胁迫 | [ |
| 其他 | CsYUC8/9、CsYUC10b | 高温、低温、盐胁迫 | [ | ||
| FMOGS-OXs | 拟南芥 | FMOGS-OX2-7 | 干旱、低温、盐、渗透胁迫及激素处理 | [ | |
| 其他 | TsFMO | 盐胁迫 | [ | ||
生物胁迫 Biological stress | FMO1 | 拟南芥 | FMO1 | 系统性获得抗性(SAR)、降解病原毒素 | [ |
| 其他 | FMO | NHP合成、抑制黄酮合成并促进木质素积累 | [ | ||
| YUCCAs | 其他 | Bs3 | 触发超敏反应 | [ | |
| FMOGS-OXs | 拟南芥 | FMOGS-OXs | GSL生物合成 | [ | |
次生代谢 Secondary metabolism | FMOGS-OXs | 拟南芥 | FMOGS-OXs | GSL生物合成 | [ |
| 其他 | FMOGS-OX2、BrrFMOGS-OX2、BrrFMOGS-OX5.1、BrrFMOGS-OX5.2、BrrFMOGS-OX6.1、BrrFMOGS-OX6.2、FMOGS-OXs、AsFMO1 | GSL生物合成、催化蒜氨酸S-氧化 | [ |
| [1] | Mitchell AJ, Weng J-K. Unleashing the synthetic power of plant oxygenases: from mechanism to application [J]. Plant Physiol, 2019, 179(3): 813-829. |
| [2] | Thodberg S, Jakobsen Neilson EH. The “green” FMOs: diversity, functionality and application of plant flavoproteins [J]. Catalysts, 2020, 10(3): 329. |
| [3] | Schlaich NL. Flavin-containing monooxygenases in plants: looking beyond detox [J]. Trends Plant Sci, 2007, 12(9): 412-418. |
| [4] | Ge CN, Gao CJ, Chen QG, et al. ESCRT-dependent vacuolar sorting and degradation of the auxin biosynthetic enzyme YUC1 flavin monooxygenase [J]. J Integr Plant Biol, 2019, 61(9): 968-973. |
| [5] | Kriechbaumer V, Wang PW, Hawes C, et al. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation [J]. Plant J, 2012, 70(2): 292-302. |
| [6] | Kim JI, Sharkhuu A, Jin JB, et al. yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes [J]. Plant Physiol, 2007, 145(3): 722-735. |
| [7] | Zhang YY, Mao QS, Ma RJ, et al. Genome-wide identification and expression analysis of the PpYUCCA gene family in weeping peach trees (Prunus persica ‘pendula’) [J]. Horticulturae, 2022, 8(10): 878. |
| [8] | Zhang K, Zhang JF, Cui C, et al. Genome-wide identification and expression profiling of the YUCCA gene family in Brassica napus [J]. Oil Crop Sci, 2022, 7(3): 103-111. |
| [9] | Li J, Kristiansen KA, Hansen BG, et al. Cellular and subcellular localization of flavin-monooxygenases involved in glucosinolate biosynthesis [J]. J Exp Bot, 2011, 62(3): 1337-1346. |
| [10] | Huijbers MME, Montersino S, Westphal AH, et al. Flavin dependent monooxygenases [J]. Arch Biochem Biophys, 2014, 544: 2-17. |
| [11] | Dai XH, Mashiguchi K, Chen QG, et al. The biochemical mechanism of auxin biosynthesis by an Arabidopsis yucca flavin -containing-containing monooxygenase [J]. J Biol Chem, 2013, 288(3): 1448-1457. |
| [12] | Yildiz I, Mantz M, Hartmann M, et al. The mobile SAR signal N-hydroxypipecolic acid induces NPR1-dependent transcriptional reprogramming and immune priming [J]. Plant Physiol, 2021, 186(3): 1679-1705. |
| [13] | Chen YC, Holmes EC, Rajniak J, et al. N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis [J]. Proc Natl Acad Sci U S A, 2018, 115(21): 4920-4929. |
| [14] | Hou XH, Liu SN, Pierri F, et al. Allelic analyses of the Arabidopsis YUC1 locus reveal residues and domains essential for the functions of YUC family of flavin monooxygenases [J]. J Integr Plant Biol, 2011, 53(1): 54-62. |
| [15] | Ahn G, Jeong SY, Khan HA, et al. FAD and NADPH binding sites of YUCCA6 are essential for chaperone activity and oxidative stress tolerance in Arabidopsis thaliana [J]. Plant Physiol Biochem, 2025, 218: 109335. |
| [16] | Jing Li BGH. Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis [J]. Plant Physiol, 2008, 148(3): 1721-1733. |
| [17] | Sun S, Bakkeren G. A bird’s-eye view: exploration of the flavin-containing monooxygenase superfamily in common wheat [J]. Front Plant Sci, 2024, 15: 1369299. |
| [18] | Gaba Y, Bhowal B, Pareek A, et al. Genomic survey of flavin monooxygenases in wild and cultivated rice provides insight into evolution and functional diversities [J]. Int J Mol Sci, 2023, 24(4): 4190. |
| [19] | Turnaev II, Gunbin KV, Suslov VV, et al. The phylogeny of class B flavoprotein monooxygenases and the origin of the YUCCA protein family [J]. Plants, 2020, 9(9): 1092. |
| [20] | Wang CY, Liu Y, Li SS, et al. Origin of plant auxin biosynthesis in charophyte algae [J]. Trends Plant Sci, 2014, 19(12): 741-743. |
| [21] | Matthes MS, Best NB, Robil JM, et al. Auxin EvoDevo: conservation and diversification of genes regulating auxin biosynthesis, transport, and signaling [J]. Mol Plant, 2019, 12(3): 298-320. |
| [22] | Hofberger JA, Lyons E, Edger PP, et al. Whole genome and tandem duplicate retention facilitated glucosinolate pathway diversification in the mustard family [J]. Genome Biol Evol, 2013, 5(11): 2155-2173. |
| [23] | Cang W, Sheng YX, Evivie ER, et al. Lineage-specific evolution of flavin-containing monooxygenases involved in aliphatic glucosinolate side-chain modification [J]. J Syst Evol, 2018, 56(2): 92-104. |
| [24] | Xie JM, Chen YR, Cai GJ, et al. Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees [J]. Nucleic Acids Res, 2023, 51(W1): W587-W592. |
| [25] | Zhang T, Li RN, Xing JL, et al. The YUCCA-auxin-WOX11 module controls crown root development in rice [J]. Front Plant Sci, 2018, 9: 523. |
| [26] | Jiang HJ, Zhai KE, Ye XF, et al. The endosperm-specific expression of YUCCA Genes enhances rice grain filling [J]. Phyton, 2022, 91(12): 2633-2648. |
| [27] | Abu-Zaitoon YM, Bennett K, Normanly J, et al. A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA [J]. Physiol Plant, 2012, 146(4): 487-499. |
| [28] | Meng Q, Zhang RL, Wang YN, et al. Genome-wide characterization and haplotypic variation analysis of the YUC gene family in foxtail millet (Setaria italica) [J]. Int J Mol Sci, 2023, 24(21): 15637. |
| [29] | Gallavotti A, Barazesh S, Malcomber S, et al. sparse inflorescence1encodes a monocot-specificYUCCA-like gene required for vegetative and reproductive development in maize [J]. Proc Natl Acad Sci U S A, 2008, 105(39): 15196-15201. |
| [30] | Henning PM, Shore JS, McCubbin AG. The S-gene YUC6 pleiotropically determines male mating type and pollen size in heterostylous Turnera (Passifloraceae): a novel neofunctionalization of the YUCCA gene family [J]. Plants, 2022, 11(19): 2640. |
| [31] | Zhu Q, Lu YC, Xiong JL, et al. Development of a stable genetic transformation system in Erigeron breviscapus: a case study with EbYUC2 in relation to leaf number and flowering time [J]. Planta, 2024, 259(5): 98. |
| [32] | Liu H, Xie WF, Zhang L, et al. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry [J]. J Integr Plant Biol, 2014, 56(4): 350-363. |
| [33] | Tatsuki M, Soeno K, Shimada Y, et al. Insertion of a transposon-like sequence in the 5'-flanking region of the YUCCA gene causes the stony hard phenotype [J]. Plant J, 2018, 96(4): 815-827. |
| [34] | Li SN, Wang CH, Zhou XY, et al. The curvature of cucumber fruits is associated with spatial variation in auxin accumulation and expression of a YUCCA biosynthesis gene [J]. Hortic Res, 2020, 7: 135. |
| [35] | Zheng L, Zhang L, Duan K, et al. YUCCA type auxin biosynthesis genes encoding flavin monooxygenases in melon: Genome-wide identification and developmental expression analysis [J]. S Afr N J Bot, 2016, 102: 142-152. |
| [36] | Müller-Moulé P, Nozue K, Pytlak ML, et al. YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance [J]. PeerJ, 2016, 4: e2574. |
| [37] | Cheng YF, Dai XH, Zhao YD. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation inArabidopsis [J]. Plant Cell, 2007, 19(8): 2430-2439. |
| [38] | Woodward C, Bemis SM, Hill EJ, et al. Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases [J]. Plant Physiol, 2005, 139(1): 192-203. |
| [39] | Chen PY, Umeda M. DNA double-strand breaks induce the expression of flavin-containing monooxygenase and reduce root meristem size in Arabidopsis thaliana [J]. Genes Cells, 2015, 20(8): 636-646. |
| [40] | Zhao P, Ma XY, Zhang RZ, et al. Integration of genome-wide association study, linkage analysis, and population transcriptome analysis to reveal the TaFMO1-5B modulating seminal root growth in bread wheat [J]. Plant J, 2023, 116(5): 1385-1400. |
| [41] | Catalá R, López-Cobollo R, Berbís MÁ, et al. Trimethylamine N-oxide is a new plant molecule that promotes abiotic stress tolerance [J]. Sci Adv, 2021, 7(21): eabd9296. |
| [42] | Cao HY, Liu RN, Zhang JH, et al. Improving sulforaphane content in transgenic broccoli plants by overexpressing MAM1, FMOGS-OX2, and Myrosinase [J]. Plant Cell Tissue Organ Cult, 2021, 146(3): 461-471. |
| [43] | Wang LL, Zhou YL, Ding Y, et al. Novel flavin-containing monooxygenase protein FMO1 interacts with CAT2 to negatively regulate drought tolerance through ROS homeostasis and ABA signaling pathway in tomato [J]. Hortic Res, 2023, 10(4): uhad037. |
| [44] | Cha JY, Kim WY, Kang SB, et al. A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis [J]. Nat Commun, 2015, 6: 8041. |
| [45] | Kim JI, Baek D, Park HC, et al. Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit [J]. Mol Plant, 2013, 6(2): 337-349. |
| [46] | Ke QB, Wang Z, Ji CY, et al. Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress [J]. Plant Physiol Biochem, 2015, 94: 19-27. |
| [47] | Nicolas-Espinosa J, Garcia-Ibañez P, Lopez-Zaplana A, et al. Confronting secondary metabolites with water uptake and transport in plants under abiotic stress [J]. Int J Mol Sci, 2023, 24(3): 2826. |
| [48] | Zhang TY, Liu R, Zheng JY, et al. Insights into glucosinolate accumulation and metabolic pathways in Isatis indigotica Fort [J]. BMC Plant Biol, 2022, 22(1): 78. |
| [49] | Sun JQ, Qi LL, Li YN, et al. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth [J]. PLoS Genet, 2012, 8(3): e1002594. |
| [50] | Yan SS, Che G, Ding L, et al. Different cucumber CsYUC genes regulate response to abiotic stresses and flower development [J]. Sci Rep, 2016, 6: 20760. |
| [51] | Ye X, Kang BG, Osburn LD, et al. Identification of the flavin-dependent monooxygenase-encoding YUCCA gene family in Populustrichocarpa and their expression in vegetative tissues and in response to hormone and environmental stresses [J]. Plant Cell Tissue Organ Cult, 2009, 97(3): 271-283. |
| [52] | Gao Q, Gao F, Wang RC, et al. Molecular cloning, expression, and polyclonal antibody production of a novel flavin-containing monooxygenase from Thellungiella Halophila [J]. Plant Mol Biol Report, 2009, 27(1): 94-101. |
| [53] | Zhao HY, Li D, Liu YQ, et al. Flavin-containing monooxygenases FMOGS-OXs integrate flowering transition and salt tolerance in Arabidopsis thaliana [J]. Physiol Plant, 2024, 176(2): e14287. |
| [54] | Czarnocka W, Fichman Y, Bernacki M, et al. FMO1 is involved in excess light stress-induced signal transduction and cell death signaling [J]. Cells, 2020, 9(10): 2163. |
| [55] | Cha JY, Jeong SY, Ahn G, et al. The thiol-reductase activity of YUCCA6 enhances nickel heavy metal stress tolerance in Arabidopsis [J]. Front Plant Sci, 2022, 13: 1007542. |
| [56] | Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana [J]. J Exp Bot, 2004, 55(407): 2331-2341. |
| [57] | Li YM, Li R, Sawada Y, et al. Abscisic acid-mediated induction of FLAVIN-CONTAINING MONOOXYGENASE 2 leads to reduced accumulation of methylthioalkyl glucosinolates in Arabidopsis thaliana [J]. Plant Sci, 2021, 303: 110764. |
| [58] | Kong WW, Li J, Yu QY, et al. Two novel flavin-containing monooxygenases involved in biosynthesis of aliphatic glucosinolates [J]. Front Plant Sci, 2016, 7: 1292. |
| [59] | Tatiana E Mishina JZ. The Arabidopsis flavin -dependent-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance [J]. Plant Physiol, 2006, 141(4): 1666-1675. |
| [60] | Hartmann M, Zeier T, Bernsdorff F, et al. Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity [J]. Cell, 2018, 173(2): 456-469.e16. |
| [61] | Koch M, Vorwerk S, Masur C, et al. A role for a flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis [J]. Plant J, 2006, 47(4): 629-639. |
| [62] | Bartsch M, Gobbato E, Bednarek P, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7 [J]. Plant Cell, 2006, 18(4): 1038-1051. |
| [63] | Ling YM, Xiong XP, Yang WL, et al. Comparative analysis of transcriptomics and metabolomics reveals defense mechanisms in melon cultivars against pseudoperonospora cubensis infection [J]. Int J Mol Sci, 2023, 24(24): 17552. |
| [64] | Christina Krönauer JK. Cell death triggered by the YUCCA-like Bs3 protein coincides with accumulation of salicylic acid and pipecolic acid but not of indole-3-acetic acid [J]. Plant Physiol, 2019, 180(3): 1647-1659. |
| [65] | Kliebenstein DJ, Kroymann J, Mitchell-Olds T. The glucosinolate-myrosinase system in an ecological and evolutionary context [J]. Curr Opin Plant Biol, 2005, 8(3): 264-271. |
| [66] | Daniel J Kliebenstein JK. Genetic control of natural variation in Arabidopsis glucosinolate accumulation [J]. Plant Physiol, 2001, 126(2): 811-825. |
| [67] | Geiselhardt S, Yoneya K, Blenn B, et al. Egg laying of cabbage white butterfly (Pieris brassicae) on Arabidopsis thaliana affects subsequent performance of the larvae [J]. PLoS One, 2013, 8(3): e59661. |
| [68] | Hopkins RJ, van Dam NM, van Loon JJA. Role of glucosinolates in insect-plant relationships and multitrophic interactions [J]. Annu Rev Entomol, 2009, 54: 57-83. |
| [69] | Calmes B, N’Guyen G, Dumur J, et al. Glucosinolate-derived isothiocyanates impact mitochondrial function in fungal cells and elicit an oxidative stress response necessary for growth recovery [J]. Front Plant Sci, 2015, 6: 414. |
| [70] | Michael Dalgaard Mikkelsen BLP. Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways [J]. Plant Physiol, 2003, 131(1): 298-308. |
| [71] | Hansen BG, Kliebenstein DJ, Halkier BA. Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis [J]. Plant J, 2007, 50(5): 902-910. |
| [72] | Li ZS, Liu YM, Li LY, et al. Transcriptome reveals the gene expression patterns of sulforaphane metabolism in broccoli florets [J]. PLoS One, 2019, 14(3): e0213902. |
| [73] | Li ZS, Liu GM, He HJ, et al. Effects of nanocarbon solution treatment on the nutrients and glucosinolate metabolism in broccoli [J]. Food Chem, 2022, 15: 100429. |
| [74] | Lee YS, Ku K-M, Becker TM, et al. Chemopreventive glucosinolate accumulation in various broccoli and collard tissues: Microfluidic-based targeted transcriptomics for by-product valorization [J]. PLoS One, 2017, 12(9): e0185112. |
| [75] | Yang Y, Hu Y, Yue YL, et al. Expression profiles of glucosinolate biosynthetic genes in turnip (Brassica rapa var. Rapa) at different developmental stages and effect of transformed flavin-containing monooxygenase genes on hairy root glucosinolate content [J]. J Sci Food Agric, 2020, 100(3): 1064-1071. |
| [76] | Zhao YJ, Chen ZY, Chen JX, et al. Comparative transcriptomic analyses of glucosinolate metabolic genes during the formation of Chinese kale seeds [J]. BMC Plant Biol, 2021, 21(1): 394. |
| [77] | Yoshimoto N, Onuma M, Mizuno S, et al. Identification of a flavin-containing S-oxygenating monooxygenase involved in alliin biosynthesis in garlic [J]. Plant J, 2015, 83(6): 941-951. |
| [78] | Grubb CD, Abel S. Glucosinolate metabolism and its control [J]. Trends Plant Sci, 2006, 11(2): 89-100. |
| [1] | 龙林茜, 曾银萍, 王茜, 邓玉萍, 葛敏茜, 陈彦灼, 李鑫娟, 杨军, 邹建. 向日葵GH3基因家族鉴定及其在花发育中的功能分析[J]. 生物技术通报, 2026, 42(1): 125-138. |
| [2] | 贺启琛, 杨扬, 阿丽亚·外力, 唐新月, 李忠喜, 陈永坤, 陈凌娜. 薰衣草CuAO基因家族特征及LaCuAO1降解生物胺功能研究[J]. 生物技术通报, 2026, 42(1): 114-124. |
| [3] | 杨丹, 靳雅荣, 毛春力, 王碧娴, 张雅宁, 杨智怡, 周芷瑶, 杨锐鸣, 范恒睿, 黄琳凯, 严海东. 象草C2H2基因家族鉴定及表达分析[J]. 生物技术通报, 2026, 42(1): 251-261. |
| [4] | 李健斌, 侯家娥, 李雷林, 艾明涛, 刘天泰, 崔秀明, 杨千. 三七脂氧合酶的全基因组鉴定及其对茉莉酸甲酯和创伤的响应[J]. 生物技术通报, 2026, 42(1): 218-229. |
| [5] | 李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158. |
| [6] | 黄国栋, 邓宇星, 程宏伟, 但焱南, 周会汶, 吴兰花. 大豆ZIP基因家族鉴定及响应铝胁迫的表达分析[J]. 生物技术通报, 2025, 41(9): 71-81. |
| [7] | 程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255. |
| [8] | 程雪, 付颖, 柴晓娇, 王红艳, 邓欣. 谷子LHC基因家族鉴定及非生物胁迫表达分析[J]. 生物技术通报, 2025, 41(8): 102-114. |
| [9] | 化文平, 刘菲, 浩佳欣, 陈尘. 丹参ADH基因家族的鉴定与表达模式分析[J]. 生物技术通报, 2025, 41(8): 211-219. |
| [10] | 腊贵晓, 赵玉龙, 代丹丹, 余永亮, 郭红霞, 史贵霞, 贾慧, 杨铁钢. 红花质膜H+-ATPase基因家族成员鉴定及响应低氮低磷胁迫的表达分析[J]. 生物技术通报, 2025, 41(8): 220-233. |
| [11] | 黄诗宇, 田姗姗, 杨天为, 高曼熔, 张尚文. 赤苍藤WRI1基因家族的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(8): 242-254. |
| [12] | 任睿斌, 司二静, 万广有, 汪军成, 姚立蓉, 张宏, 马小乐, 李葆春, 王化俊, 孟亚雄. 大麦条纹病菌GH17基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 146-154. |
| [13] | 张泽, 杨秀丽, 宁东贤. 花生4CL基因家族鉴定及对干旱与盐胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 117-127. |
| [14] | 李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138. |
| [15] | 王从欢, 伍国强, 魏明. 植物CBL调控逆境胁迫响应的作用机制[J]. 生物技术通报, 2025, 41(7): 1-16. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||