








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (5): 56-66.doi: 10.13560/j.cnki.biotech.bull.1985.2020-0919
Previous Articles Next Articles
QIU Xiao-yu1( ), LIU Zuo-hua1,2, QI Ren-li1,3(
), LIU Zuo-hua1,2, QI Ren-li1,3( )
)
Received:2020-07-22
Online:2021-05-26
Published:2021-06-11
Contact:
QI Ren-li
E-mail:1129363375@qq.com;qirenli1982@163.com
QIU Xiao-yu, LIU Zuo-hua, QI Ren-li. Differences in Early Fat Development and Gene Transcription Expression in the Adipose Tissues of Piglets with and Without Gut Microbiota[J]. Biotechnology Bulletin, 2021, 37(5): 56-66.

Fig. 1 Difference of fat deposition between GF pigs and normal pigs A: He staining of fat tissue of GF pigs and normal pigs. B: Back-fat of GF pigs and Normal pigs. C: Diameter of fat cells in fat tissue of GF pigs and Normal pigs. *** P<0.001;**** P<0.0001

Fig.2 Differences in fat synthesis regulatory factors and adipocytokines between GF pigs and normal pigs A: The differential expression of FABP4, PPAR γ, ACC and FAS in adipose tissue of GF and Normal piglets. B: The differential secretion of adiponectin. C: The differential secretion of adiponectin leptin. * P<0.05;** P<0.01
| 样品名 Sample name | 总序列数 Raw reads | 干净序列数 Clean reads | 总对比到的序列数 Total mapped reads | 多重对比的序列数 Multiple mapped reads | 唯一对比的序列数 Uniquely mapped reads |
|---|---|---|---|---|---|
| Normal A1 | 60 328 616 | 59 694 488 | 50 892 462(85.25%) | 4 145 489(6.94%) | 46 746 973(78.31%) |
| Normal A2 | 53 878 956 | 53 400 340 | 45 855 826(85.87%) | 4 209 183(7.88%) | 41 646 643(77.99%) |
| Normal A3 | 65 393 020 | 64 761 370 | 55 361 124(85.48%) | 5 082 167(7.85%) | 50 278 957(77.64%) |
| GF A1 | 48 721 914 | 48 260 760 | 41 315 919(85.61%) | 2 945 880(6.10%) | 38 370 039(79.51%) |
| GF A2 | 54 262 278 | 53 749 144 | 46 093 576(85.76%) | 3 467 323(6.45%) | 42 626 253(79.31%) |
| GF A3 | 57 431 858 | 56 874 014 | 48 683 278(85.60%) | 3 484 909(6.13%) | 45 198 369(79.47%) |
Table 1 Statistical list of mapping to genome
| 样品名 Sample name | 总序列数 Raw reads | 干净序列数 Clean reads | 总对比到的序列数 Total mapped reads | 多重对比的序列数 Multiple mapped reads | 唯一对比的序列数 Uniquely mapped reads |
|---|---|---|---|---|---|
| Normal A1 | 60 328 616 | 59 694 488 | 50 892 462(85.25%) | 4 145 489(6.94%) | 46 746 973(78.31%) |
| Normal A2 | 53 878 956 | 53 400 340 | 45 855 826(85.87%) | 4 209 183(7.88%) | 41 646 643(77.99%) |
| Normal A3 | 65 393 020 | 64 761 370 | 55 361 124(85.48%) | 5 082 167(7.85%) | 50 278 957(77.64%) |
| GF A1 | 48 721 914 | 48 260 760 | 41 315 919(85.61%) | 2 945 880(6.10%) | 38 370 039(79.51%) |
| GF A2 | 54 262 278 | 53 749 144 | 46 093 576(85.76%) | 3 467 323(6.45%) | 42 626 253(79.31%) |
| GF A3 | 57 431 858 | 56 874 014 | 48 683 278(85.60%) | 3 484 909(6.13%) | 45 198 369(79.47%) |

Fig.5 Differentially expressed genes A:The Veen map of differentially expressed gene. B:The volcano map of differentially expressed gene. C:The clustering map of differentially expressed gene
| 基因名称Gene Name | log2FC | 变化Change | 功能Function |
|---|---|---|---|
| TNS4 | 4.756 | Up | Function unknown |
| TMP | 4.587 | Up | Function unknown |
| NRIP3 | 4.305 | Up | Proteolysis,aspartic-type endopeptidase activity |
| MAPKAPK | 4.088 | Up | ATP binding,protein kinase activity |
| PRG4 | 4.076 | Up | Receptor-mediated endocytosis |
| IRF5 | 4.051 | Up | Function unknown |
| IRF6 | 4.007 | Up | Keratinocyte proliferation |
| ITIH3 | 3.668 | Up | Hyaluronan metabolic process |
| 3,5,3’,5’- tetraiodothyronine | 3.566 | Up | Hormone biosynthetic process |
| Phenylalanine 4-monooxygenase | 3.55 | Up | Amino acid transport and metabolism,oxidoreductase activity |
| C4BPA | -7.537 | Down | Positive regulation of protein catabolic |
| SQRDL | -6.269 | Down | Oxidoreductase activity |
| PIK3C2G | -4.847 | Down | Phosphatidylinositol 3-kinase(PI3K)complex |
| KCNIP2 | -4.293 | Down | Cell cycle control,myosin light chain |
| LIPG | -4.149 | Down | Lipid metabolic,high-density lipoprotein particle remodeling |
| MAP6 | -3.894 | Down | Lysosome localization |
| CES1 | -3.625 | Down | Lipid transport and metabolism,Carboxylesterase |
| CYP1A1 | -3.588 | Down | Steroid metabolic,Steroid hormone biosynthesis |
| SucCβ | -3.498 | Down | Fatty acid biosynthetic process,acetyl-CoA biosynthetic process |
| LRFN2 | -3.452 | Down | Function unknown |
Table 2 Top 10 up or down expressed genes
| 基因名称Gene Name | log2FC | 变化Change | 功能Function |
|---|---|---|---|
| TNS4 | 4.756 | Up | Function unknown |
| TMP | 4.587 | Up | Function unknown |
| NRIP3 | 4.305 | Up | Proteolysis,aspartic-type endopeptidase activity |
| MAPKAPK | 4.088 | Up | ATP binding,protein kinase activity |
| PRG4 | 4.076 | Up | Receptor-mediated endocytosis |
| IRF5 | 4.051 | Up | Function unknown |
| IRF6 | 4.007 | Up | Keratinocyte proliferation |
| ITIH3 | 3.668 | Up | Hyaluronan metabolic process |
| 3,5,3’,5’- tetraiodothyronine | 3.566 | Up | Hormone biosynthetic process |
| Phenylalanine 4-monooxygenase | 3.55 | Up | Amino acid transport and metabolism,oxidoreductase activity |
| C4BPA | -7.537 | Down | Positive regulation of protein catabolic |
| SQRDL | -6.269 | Down | Oxidoreductase activity |
| PIK3C2G | -4.847 | Down | Phosphatidylinositol 3-kinase(PI3K)complex |
| KCNIP2 | -4.293 | Down | Cell cycle control,myosin light chain |
| LIPG | -4.149 | Down | Lipid metabolic,high-density lipoprotein particle remodeling |
| MAP6 | -3.894 | Down | Lysosome localization |
| CES1 | -3.625 | Down | Lipid transport and metabolism,Carboxylesterase |
| CYP1A1 | -3.588 | Down | Steroid metabolic,Steroid hormone biosynthesis |
| SucCβ | -3.498 | Down | Fatty acid biosynthetic process,acetyl-CoA biosynthetic process |
| LRFN2 | -3.452 | Down | Function unknown |
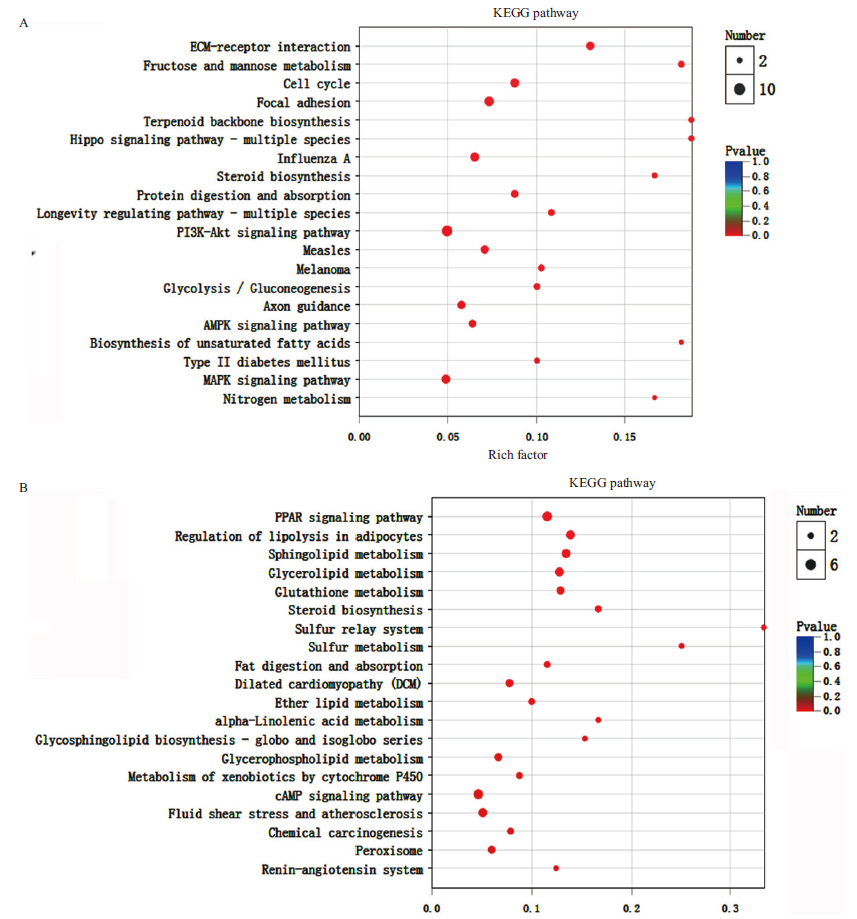
Fig.7 KEGG enrichment analysis of the differentially expressed genes A: KEGG analysis of the down-regulated genes. B:KEGG analysis of the up-regulated genes
| [1] |
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome[J]. Nature, 2012,486(7402):207-214.
doi: 10.1038/nature11234 URL |
| [2] |
Kim HB, Isaacson RE. The pig gut microbial diversity:Understanding the pig gut microbial ecology through the next generation high throughput sequencing[J]. Veterinary Microbiology, 2015,177(3):242-251.
doi: 10.1016/j.vetmic.2015.03.014 URL |
| [3] | 唐义梅. 梅山猪粪便微生物移植对长×大后备母猪卵泡发育的影响[D]. 武汉:华中农业大学, 2019. |
| Tang YM. Effect of fecal microbiota transplantation of Meishan pig on follicle development in Landrace × Yorkshire gilts[D]. Wuhan:Huazhong Agricultural University, 2019. | |
| [4] |
Ejtahed HS, Hasani-Ranjbar S. Neuromodulatory effect of microbiome on gut-brain axis;new target for obesity drugs[J]. Journal of Diabetes and Metabolic Disorders, 2019,18(1):263-265.
doi: 10.1007/s40200-019-00384-4 URL |
| [5] |
Fukui H. Role of gut dysbiosis in liver diseases:what have we learned so far?[J]. Diseases, 2019,7(4):58-65.
doi: 10.3390/diseases7040058 URL |
| [6] |
Gao J, Xu K, Liu H, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism[J]. Frontiers in Cellular and Infection Microbiology, 2018,8(13):16-24.
doi: 10.3389/fcimb.2018.00016 URL |
| [7] |
Boulangé CL, Neves AL, Chilloux J, et al. Impact of the gut microbiota on inflammation, obesity, and metabolic disease[J]. Genome Medicine, 2016,8(1):42-58.
doi: 10.1186/s13073-016-0303-2 pmid: 27098727 |
| [8] | Ebrahimzadeh LH, Sanaie S, Sadeghpour HF, et al. From role of gut microbiota to microbial-based therapies in type 2-diabetes[J]. Infection, Genetics and Evolution, 2020,81(1):153-167. |
| [9] |
Miraglia F, Colla E. Microbiome, Parkinson’s disease and molecular mimicry[J]. Cells, 2019,8(3):222-230.
doi: 10.3390/cells8030222 URL |
| [10] |
Heym N, Heasman BC, Hunter K, et al. The role of microbiota and inflammation in self-judgement and empathy:implications for understanding the brain-gut-microbiome axis in depression[J]. Psychopharmacology, 2019,236(5):1459-1470.
doi: 10.1007/s00213-019-05230-2 pmid: 30955108 |
| [11] | 张学娇. YAP蛋白对脂肪功能的影响及其在肥胖相关代谢性疾病中的作用[D]. 天津:天津医科大学, 2017. |
| Zhang XJ. Role of YAP in adipose tissue function and the obesity related metabolic diseases[D]. Tianjin:Tianjin Medical University, 2017. | |
| [12] | 张震. 鼠李糖乳酸杆菌LGG来源胞外多糖抑制动物脂肪生成的研究[D]. 北京:中国农业科学院, 2017. |
| Zhang Z. Isolated exopolysaccharides from Lactobacillus rhamnosus GG inhibit adipogenesis in animals[D]. Beijing:Chinese Academy of Agricultural Sciences, 2017. | |
| [13] |
Cani PD. Metabolism in 2013:The gut microbiota manages host metabolism[J]. Nature Reviews Endocrinology, 2014,10(2):74-76.
doi: 10.1038/nrendo.2013.240 URL |
| [14] | Campbell JH, Foster CM, Vishnivetskaya T, et al. Host genetic and environmental effects on mouse intestinal microbiota[J]. Multidisciplinary Journal of Microbial Ecology, 2012,6(1):2033-2044. |
| [15] |
Dhakal S, Wang L, Antony L, et al. Amish(Rural)vs. non-Amish(Urban)infant fecal microbiotas are highly diverse and their transplantation lead to differences in mucosal immune maturation in a humanized germfree piglet model[J]. Frontiers in Immunology, 2019,10(1):1509-1517.
doi: 10.3389/fimmu.2019.01509 URL |
| [16] | Potockova H, Sinkorova J, Karova K, Sinkora M. The distribution of lymphoid cells in the small intestine of germ-free and conventional piglets[J]. Devlopmental and Comparative Immunology, 2015,51(1):99-107. |
| [17] |
Sinkora M, Butler JE, Lager KM, et al. The comparative profile of lymphoid cells and the T and B cell spectratype of germ-free piglets infected with viruses SIV, PRRSV or PCV2[J]. Veterinary Research, 2014,45(1):91-105.
doi: 10.1186/s13567-014-0091-x URL |
| [18] | Xiao L, Estellé J, Kiilerich P, et al. A reference gene catalogue of the pig gut microbiome[J]. Nature Microbiology, 2016,1(1):179-184. |
| [19] | 孙静, 杜蕾, 丁玉春, 等. 无菌猪的制备与微生物质量控制[J]. 中国实验动物学报, 2017,25(6):699-702. |
| Sun J, Du L, Ding YC, et al. Breeding and microbiological quality control of germ-free pigs[J]. Acta Laboratorium Animalis Scientia Sinica, 2017,25(6):699-702. | |
| [20] |
Hu J, Lin S, Zheng B, et al. Short-chain fatty acids in control of energy metabolism[J]. Critical Reviews in Food Science and Nutrition, 2018,58(8):1243-1249.
doi: 10.1080/10408398.2016.1245650 URL |
| [21] |
Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage[J]. PNAS, 2004,101(44):15718-15723.
doi: 10.1073/pnas.0407076101 URL |
| [22] |
Tilg H, Moschen AR. Microbiota and diabetes:an evolving relationship[J]. Gut, 2014,63(9):1513-1521.
doi: 10.1136/gutjnl-2014-306928 URL |
| [23] |
Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins[J]. Nature, 2009,457(28):480-484.
doi: 10.1038/nature07540 URL |
| [24] | Toda G, Soeda K, Okazaki Y, et al. Insulin- and lipopolysaccharide-mediated signaling in adipose tissue macrophages regulates postprandial glycemia through Akt-mTOR activation[J]. Moleculer Cell, 2020,79(1):43-53. |
| [25] |
Ding XM, Li DD, Bai SP, et al. Effect of dietary xylooligosaccharides on intestinal characteristics, gut microbiota, cecal short-chain fatty acids, and plasma immune parameters of laying hens[J]. Poultry Science, 2018,97(3):874-881.
doi: 10.3382/ps/pex372 pmid: 29294100 |
| [26] | Massier L, Chakaroun R, Tabei S, et al. Adipose tissue derived bacteria are associated with inflammation in obesity and type 2 diabetes[J]. Gut, 2020,4(2):21-41. |
| [27] |
Russo R, Cristiano C, Avagliano C, et al. Gut-brain axis:role of lipids in the regulation of inflammation, pain and CNS diseases[J]. Current Medicinal Chemistry, 2018,25(32):3930-3952.
doi: 10.2174/0929867324666170216113756 URL |
| [28] |
Muller PA, Schneeberger M, Matheis F, et al. Microbiota modulate sympathetic neurons via a gut-brain circuit[J]. Nature, 2020,583(7823):441-446.
doi: 10.1038/s41586-020-2474-7 URL |
| [29] |
Perry RJ, Peng L, Barry N. A, et al. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome[J]. Nature, 2016,534(7606):213-217.
doi: 10.1038/nature18309 URL |
| [30] |
Brown AJ, Goldsworthy SM, Barnes, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids[J]. Journal of Biological Chemistry, 2003,278(13):11312-11319.
doi: 10.1074/jbc.M211609200 URL |
| [31] |
Xiong Y, Miyamoto N, Shibata K, et al. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41[J]. PNAS, 2004,101(4):1045-1050.
doi: 10.1073/pnas.2637002100 URL |
| [32] | Gaestel M. MAPK-Activated protein kinases(MKs):novel insights and challenges[J]. Frontiers in Cell and Development Biology, 2016,3(8):88-96. |
| [33] |
Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases[J]. Microbiology and Molecular Biology Reviews, 2011,75(1):50-83.
doi: 10.1128/MMBR.00031-10 URL |
| [34] | Sidarala V, Kowluru A. The regulatory roles of Mitogen-Activated Protein Kinase(MAPK)pathways in health and diabetes:lessons learned from the pancreatic β-Cell[J]. Metabolic and Immune Drug Discovery, 2017,10(2):76-84. |
| [35] | Wickramasekara RN, Morrill S, Farhat Y, et al. Glutathione and Inter-α-trypsin inhibitor heavy chain 3(Itih3)mRNA levels in nicotine-treated Cd44 knockout mice[J]. Toxicology Reports, 2018,22(5):759-764. |
| [36] |
Bost F, Diarra-Mehrpour M, Martin JP. Inter-alpha-trypsin inhibitor proteoglycan family--a group of proteins binding and stabilizing the extracellular matrix[J]. European Journal of Biochemisty, 1998,252(3):339-346.
doi: 10.1046/j.1432-1327.1998.2520339.x URL |
| [37] |
Lichter-Konecki U, Vockley J. Phenylketonuria:current treatments and future developments[J]. Drugs, 2019,79(5):495-500.
doi: 10.1007/s40265-019-01079-z pmid: 30864096 |
| [38] |
Braccini L, Ciraolo E, Campa CC, et al. PI3K-C2γ is a Rab5 effector selectively controlling endosomal Akt2 activation downstream of insulin signalling[J]. Nature Communications, 2015,6(1):7400-7415.
doi: 10.1038/ncomms8400 URL |
| [39] | Yu JE, Han SY, Wolfson B, et al. The role of endothelial lipase in lipid metabolism, inflammation, and cancer[J]. Histology and Histopatholoy, 2018,33(1):1-10. |
| [40] |
Lian J, Nelson R, Lehner R. Carboxylesterases in lipid metaboli-sm:from mouse to human[J]. Protein Cell, 2018,9(1):178-195.
doi: 10.1007/s13238-017-0437-z URL |
| [41] | Wang D, Zou L, Jin Q, et al. Human carboxylesterases:a compr-ehensive review[J]. Acta Pharm Sin B, 2018,8(5):9-22. |
| [42] | Zhang WY, Wang H, Qi S, et al. CYP1A1 relieves lipopolysacc-haride-induced inflammatory responses in Bovine mammary epithelial cells[J]. Mediators Inflamm, 2018,1(3):1-10. |
| [43] |
Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage[J]. PNAS, 2004,101(44):15718-15723.
doi: 10.1073/pnas.0407076101 URL |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | LOU Hui, ZHU Jin-cheng, YANG Yang, ZHANG Wei. Effects of Root Exudates in Resistant and Susceptible Varieties of Cotton on the Growths and Gene Expressions of Fusarium oxysporum [J]. Biotechnology Bulletin, 2023, 39(9): 156-167. |
| [3] | LIU Yu-ling, WANG Meng-yao, SUN Qi, MA Li-hua, ZHU Xin-xia. Effect of RD29A Promoter on the Stress Resistance of Transgenic Tobacco with SikCDPK1 Gene from Saussurea involucrata [J]. Biotechnology Bulletin, 2023, 39(9): 168-175. |
| [4] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [5] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [6] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [7] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [8] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [9] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [10] | HE Yu-hang, HU Tao, WU Zhen, HE Yu, CHENG An-chun, CHEN Shun. Establishment of YFV17D Non-infectious Reporter Replicon and Pseudovirus Packaging System [J]. Biotechnology Bulletin, 2023, 39(8): 165-172. |
| [11] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [12] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [13] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [14] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [15] | ZHANG Dao-lei, GAN Yu-jun, LE Liang, PU Li. Epigenetic Regulation of Yield-related Traits in Maize and Epibreeding [J]. Biotechnology Bulletin, 2023, 39(8): 31-42. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||