








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (5): 67-75.doi: 10.13560/j.cnki.biotech.bull.1985.2020-0963
Previous Articles Next Articles
XIE Shao-yi( ), JIANG Lu-man, YANG Xiao-feng, ZHANG Xian-yu, WU Zhen-fang, LI Zi-cong(
), JIANG Lu-man, YANG Xiao-feng, ZHANG Xian-yu, WU Zhen-fang, LI Zi-cong( )
)
Received:2020-08-03
Online:2021-05-26
Published:2021-06-11
Contact:
LI Zi-cong
E-mail:593293238@qq.com;lizicong@scau.edu.cn
XIE Shao-yi, JIANG Lu-man, YANG Xiao-feng, ZHANG Xian-yu, WU Zhen-fang, LI Zi-cong. Construction and Verification of CRISPR/Cas9 System Expression Vector for Mouse X Chromosome Cutting[J]. Biotechnology Bulletin, 2021, 37(5): 67-75.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物长度 Product size/bp |
|---|---|---|
| X1-F | TGAGTACTTGGTTAGGGTGAGC | 1 212 |
| X1-R | TGGAAGTCTCTGGCATA | |
| X3-F | AAGGTTCCCAATGGTCACAGG | 1 383 |
| X3-R | ACCATACCATGGTTTTCCCCA |
Table 1 Target fragment amplification primers
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物长度 Product size/bp |
|---|---|---|
| X1-F | TGAGTACTTGGTTAGGGTGAGC | 1 212 |
| X1-R | TGGAAGTCTCTGGCATA | |
| X3-F | AAGGTTCCCAATGGTCACAGG | 1 383 |
| X3-R | ACCATACCATGGTTTTCCCCA |
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物长度 Product size/bp |
|---|---|---|
| sgRNA X1-F | TAATACGACTCACTATAGGGG- CTTGGTTAGGGTGAGTGCT | 120 |
| sgRNA X1-R | AAAAGCACCGACTCGGTGCC | |
| sgRNA X3-F | TAATACGACTCACTATAGGG- TAAGTGCTGTGTGCTGCTAC | 120 |
| sgRNA X3-R | AAAAGCACCGACTCGGTGCC |
Table 2 Primers for amplification of sgRNA transcription template in vitro
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物长度 Product size/bp |
|---|---|---|
| sgRNA X1-F | TAATACGACTCACTATAGGGG- CTTGGTTAGGGTGAGTGCT | 120 |
| sgRNA X1-R | AAAAGCACCGACTCGGTGCC | |
| sgRNA X3-F | TAATACGACTCACTATAGGG- TAAGTGCTGTGTGCTGCTAC | 120 |
| sgRNA X3-R | AAAAGCACCGACTCGGTGCC |
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物片段 Product size/bp |
|---|---|---|
| X1 mutation-F | GCATGCTTGGTTAGGGTGAGTT | 954 |
| X1 mutation-R | GCTGGAAACCCAAGCAC |
Table 3 Primers for amplification of the X1 target site
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物片段 Product size/bp |
|---|---|---|
| X1 mutation-F | GCATGCTTGGTTAGGGTGAGTT | 954 |
| X1 mutation-R | GCTGGAAACCCAAGCAC |
| sgRNA名称 sgRNA name | 靶位点(拷贝数) Target location(copy number) | 核苷酸序列 Nucleotide sequence(5'-3') |
|---|---|---|
| sgRNA X1 | X1(72) | GCTTGGTTAGGGTGAGTGCT |
| sgRNA X3 | X3(99) | TAAGTGCTGTGTGCTGCTAC |
Table 4 Sequences of sgRNA targeting mouse X chromosome multi-copy genes
| sgRNA名称 sgRNA name | 靶位点(拷贝数) Target location(copy number) | 核苷酸序列 Nucleotide sequence(5'-3') |
|---|---|---|
| sgRNA X1 | X1(72) | GCTTGGTTAGGGTGAGTGCT |
| sgRNA X3 | X3(99) | TAAGTGCTGTGTGCTGCTAC |
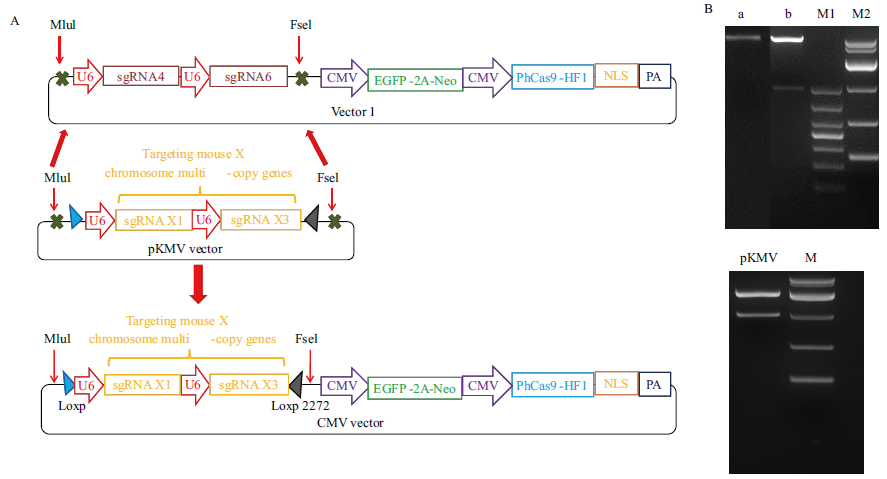
Fig.1 Construction and identification of CMV vector A: Schematic diagram of CMV carrier construction. B: Electrophoresis diagram of plasmid double digestion results synthesized by vector 1 and sgRNA fragments, M1 is 1000 DNA marker, M2 is 10000 DNA marker, A is the original vector 1, B is the digested vector 1, M is 10000 DNA marker, and pKMV is a plasmid synthesizing sgRNA fragment
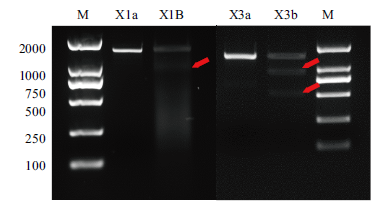
Fig. 2 Electrophoresis diagram of verifying the efficiency of sgRNA X1 and sgRNA X3 mediated Cas9 protein cleaving target sequence in vitro a is the uncut sgRNA target sequence fragment, b is the cut sgRNA target sequence fragment, arrow indicates the cut fragment, and M is 2000 DNA marker

Fig.3 Comparison results of the number of fluorescent cells in MLTC-1 cells at different time after transfecting with CMV vector and empty vector A: Fluorescence renderings at different time points after male cells were transfected with CMV vector and empty vector. B: Proportion of green fluorescent cells at different time points after vector transfection

Fig. 4 Sequencing results of the X1 target site of the monoclonal cell mass of MLTC-1 cells after CMV vector transfection Target sites are marked in the red box, * is the matching base, and the missing space is the mutation base
| 转染细胞 Transfection cell | 检测筛选获得的单克隆细胞团数 Number of monoclonal cell clusters obtained by screening was detected | 突变的单克隆细胞团数 Number of mutant monoclonal cell clusters | 突变效率 Mutation efficiency/% |
|---|---|---|---|
| MLTC-1 | 36 | 4 | 11.1 |
Table 5 Mutation efficiency of CMV vector transfected MLTC-1 cell X1 target site
| 转染细胞 Transfection cell | 检测筛选获得的单克隆细胞团数 Number of monoclonal cell clusters obtained by screening was detected | 突变的单克隆细胞团数 Number of mutant monoclonal cell clusters | 突变效率 Mutation efficiency/% |
|---|---|---|---|
| MLTC-1 | 36 | 4 | 11.1 |
| 组别 Group | 注射胚胎数Number of injected embryos | 卵裂胚数Number of cleavage embryo | 卵裂率 Cleavage rate | 囊胚数Number of blastocysts | 囊胚率 Blastocyst rate | 囊胚细胞数 Number of blastocyst cells |
|---|---|---|---|---|---|---|
| 注射 CMV 载体组 The Group of CMV vector injection | 98 | 96 | 98.0% | 57 | 58.1% a | 33.93±4.93 a |
| 注射空载体组 The Group of empty vector injection | 92 | 88 | 95.7% | 64 | 69.5% a | 44.66±6.18 b |
Table 6 Comparison of in vitro development efficiency of parthenogenetic embryos injected with CMV vector andempty vector
| 组别 Group | 注射胚胎数Number of injected embryos | 卵裂胚数Number of cleavage embryo | 卵裂率 Cleavage rate | 囊胚数Number of blastocysts | 囊胚率 Blastocyst rate | 囊胚细胞数 Number of blastocyst cells |
|---|---|---|---|---|---|---|
| 注射 CMV 载体组 The Group of CMV vector injection | 98 | 96 | 98.0% | 57 | 58.1% a | 33.93±4.93 a |
| 注射空载体组 The Group of empty vector injection | 92 | 88 | 95.7% | 64 | 69.5% a | 44.66±6.18 b |
| [1] | 李永福. 家畜性别控制技术研究进展[J]. 中国草食动物, 2003. 23(6):33-34. |
| Li YF. Progress in livestock sex control technology[J]. Chinese Herbivores, 2003. 23(6):33-34. | |
| [2] | 廖勤丰, 何航, 陈亚强, 等. 哺乳动物性别决定相关基因研究进展[J]. 家畜生态学报, 2019. 40(7):9-12. |
| Liao QF, He H, Chen YQ, et al. Advances in the study of genes related to sex determination in mammals[J]. Journal of Domestic Animal Ecology, 2019. 40(7):9-12. | |
| [3] |
Marei WFA, Khalil WA, Pushpakumara APG, et al. Polyunsaturated fatty acids influence offspring sex ratio in cows[J]. International Journal of Veterinary Science and Medicine, 2018. 6:S36-S40.
doi: 10.1016/j.ijvsm.2018.01.006 URL |
| [4] |
Song WH, Mohamed EA, Pang WK, et al. Effect of endocrine disruptors on the ratio of X and Y chromosome-bearing live spermatozoa[J]. Reproductive Toxicology, 2018. 82:10-17.
doi: 10.1016/j.reprotox.2018.09.002 URL |
| [5] | 赵红恩. 受精时阴道酸碱度变化对子代家兔性别的影响[J]. 中国优生与遗传杂志, 2015. 23(9):119. |
| Zhao HE. The effect of changes in vaginal pH during fertilization on the sex of offspring rabbits[J]. Chinese Journal of Health and Genetics, 2015. 23(9):119. | |
| [6] | 陆阳清, 张明, 卢克焕. 流式细胞仪分离精子法的研究进展[J]. 生物技术通报, 2005(3):26-30. |
| Lu YQ, Zhang M, Lu KH. Progress in flow cytometry for sperm isolation[J]. Biotechnology Bulletin, 2005(3):26-30. | |
| [7] |
Schenk JL, Suh TK, Cran DG, et al. Cryopreservation of flow-sorted bovine spermatozoa[J]. Theriogenology, 1999,52(8):1375-1391.
pmid: 10735083 |
| [8] | 王海浪, 薛建华, 吕小青, 等. 流式细胞仪生产牛性控冷冻精液技术要点[J]. 中国奶牛, 2014(1):53-55. |
| Wang HL, Xue JH, Lv XQ, et al. Key points of flow cytometry for bovine sex control and frozen semen production[J]. Chinese Dairy Cows, 2014(1):53-55. | |
| [9] |
Galizi R, Doyle LA, Menichelli M, et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito[J]. Nature Communications, 2014. 5(1):3977.
doi: 10.1038/ncomms4977 URL |
| [10] |
Galizi R, Hammond A, Kyrou K, et al. A CRISPR-Cas9 sex-ratio distortion system for genetic control[J]. Scientific Reports, 2016. 6(1):31139.
doi: 10.1038/srep31139 URL |
| [11] | 杨晓峰. 利用 CRISPRCas9 系统打靶 Rbmy 基因对小鼠性别比例影响的初步研究[D]. 广州:华南农业大学, 2017. |
| Yang XF. Preliminary study on the effect of CRISPRCas9 targeting Rbmy gene on the sex ratio in mice[D]. Guangzhou:South China Agricultural University, 2017. | |
| [12] |
Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, CRISPR/Cas-based methods for genome engineering[J]. Trends in Biotechnology, 2013,31(7):397-405.
doi: 10.1016/j.tibtech.2013.04.004 URL |
| [13] | 邓倩云, 常雪莹, 王斯佳, 等. 利用 CRISPR 系统介导人类 mir-505 基因位点的编辑[J]. 现代生物医学进展, 2016,16(2):257-260. |
| Deng QY, Chang XY, Wang SJ, et al. CRISPR mediated editing of human mir-505 gene loci[J]. Progress in Modern Biomedicine, 2016,16(2):257-260. | |
| [14] | 吴金青, 梅瑰, 刘志国, 等. 应用 SSA 报告载体提高 ZFN 和 CRISPR/Cas9 对猪 IGF2 基因的打靶效率[J]. 遗传, 2015,37(1):55-62. |
| Wu JQ, Mei G, Liu ZG, et al. Application of SSA reporting vector to improve the targeting efficiency of ZFN and CRISPR/Cas9 on pig IGF2 gene[J]. Hereditas, 2015,37(1):55-62. | |
| [15] | 刘燕, 任卫红, 王绿娅, 等. CRISPR/Cas9 基因编辑系统中两种 gRNA 活性检测方法比较[J]. 心肺血管病杂志, 2016,35(7):563-568. |
| Liu Y, Ren WH, Wang LY, et al. Comparison of two gRNA activity detection methods in CRISPR/Cas9 gene editing system[J]. Journal of Cardiovascular and Pulmonary Diseases, 2016,35(7):563-568. | |
| [16] |
Zuo E, Huo X, Yao X, et al. CRISPR/Cas9-mediated targeted chromosome elimination[J]. Genome Biology, 2017,18(1):224.
doi: 10.1186/s13059-017-1354-4 URL |
| [17] | 严雨倩, 周平坤. 哺乳动物细胞 DNA 非同源末端连接及其生物学意义[J]. 医学分子生物学杂志, 2006,3(1):69-72. |
| Yan YQ, Zhou PK. Non-homologous terminal connections of mammalian cell DNA and their biological significance[J]. Journal of Medical Molecular Biology, 2006,3(1):69-72. | |
| [18] |
Burma S, Chen BPC, Chen DJ. Role of non-homologous end joining(NHEJ)in maintaining genomic integrity[J]. DNA Repair, 2006,5(9):1042-1048.
doi: 10.1016/j.dnarep.2006.05.026 URL |
| [19] |
Bergs JWJ, Krawczyk PM, Borovski T, et al. Inhibition of homologous recombination by hyperthermia shunts early double strand break repair to non-homologous end-joining[J]. DNA Repair, 2013,12(1):38-45.
doi: 10.1016/j.dnarep.2012.10.008 URL |
| [20] | 蒋璐蔓, 李嵩, 杨晓峰, 等. 靶向小鼠 Rbmy 基因的 CRISPR/Cas9 系统高效切割位点筛选及其表达载体构建[J]. 畜牧与兽医, 2019,51(12):1-8. |
| Jiang LM, Li S, Yang XF, et al. Screening and expression vector construction of CRISPR/Cas9 system targeting Rbmy gene in mice[J]. Anim Husban Veter Med, 2019,51(12):1-8. | |
| [21] | 朱新产, 王宝维. 胚胎发育合子型基因激活(ZGA)的调控机制[J]. 西北农林科技大学学报:自然科学版, 2000,23(2):91-97. |
| Zhu XC, Wang BW. Regulation mechanism of zygotic gene activation(ZGA)in embryonic development[J]. Journal of Northwest A & F University:Natural Science Edition, 2000,23(2):91-97. | |
| [22] | 刘红林, 汪河海, 范必勤, 等. 乙醇激活诱导小鼠孤雌胚的发育与核型分析[J]. 畜牧与兽医, 2000,32(1):7-8. |
| Liu HL, Wang HH, Fan BQ, et al. In vitro development and karyotypes of ethanol activated parthenogenetic embryos in mouse[J]. Anim Husbn Veter Med, 2000,32(1):7-8. |
| [1] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [2] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [3] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [4] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [5] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [6] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [7] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [8] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [9] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [10] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [11] | Olalekan Amoo, HU Li-min, ZHAI Yun-gu, FAN Chu-chuan, ZHOU Yong-ming. Regulation of Shoot Branching by BRANCHED1 in Brassica napus Based on Gene Editing Technology [J]. Biotechnology Bulletin, 2022, 38(4): 97-105. |
| [12] | DING Ya-qun, DING Ning, XIE Shen-min, HUANG Meng-na, ZHANG Yu, ZHANG Qin, JIANG Li. Construction of Vps28 Knock-out Mice and Model Study of the Impact on Lactation and Immune Traits [J]. Biotechnology Bulletin, 2022, 38(3): 164-172. |
| [13] | YAN Jiong, FENG Chen-yi, GAO Xue-kun, XU Xiang, YANG Jia-min, CHEN Zhao-yang. Construction of Homozygous Plin1-knockout Mouse Model and Phenotype Analysis Based on CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2022, 38(3): 173-180. |
| [14] | LI Zhi-hao, ZHANG Ge, MO Zhi-jie, DENG Shuai-jun, LI Jia-yi, ZHANG Hai-bo, LIU Xiao-hui, LIU Hao-bao. Effects of a Xylanase-producing Bacillus cereus on the Composition and Fermented Products of Cigar Leaves [J]. Biotechnology Bulletin, 2022, 38(2): 105-112. |
| [15] | ZHONG Jing, SUN Ling-ling, ZHANG Shu, MENG Yuan, ZHI Yi-fei, TU Li-qing, XU Tian-peng, PU Li-ping, LU Yang-qing. Effect of Knocking Out the Mda5 Gene by CRISPR/Cas9 Technology on the Replication of Newcastle Disease and Infectious Bursal Virus [J]. Biotechnology Bulletin, 2022, 38(11): 90-96. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||