








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (5): 141-153.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1452
Previous Articles Next Articles
GAO Peng-fei1( ), XI Fei-hu1,2, ZHANG Ze-yu1, HU Kai-qiang1, CHEN Kai2, WEI Wen-tao1, DING Jia-zhi1, GU Lian-feng1(
), XI Fei-hu1,2, ZHANG Ze-yu1, HU Kai-qiang1, CHEN Kai2, WEI Wen-tao1, DING Jia-zhi1, GU Lian-feng1( )
)
Received:2020-11-27
Online:2021-05-26
Published:2021-06-11
Contact:
GU Lian-feng
E-mail:pfgao925@163.com;lfgu@fafu.edu.cn
GAO Peng-fei, XI Fei-hu, ZHANG Ze-yu, HU Kai-qiang, CHEN Kai, WEI Wen-tao, DING Jia-zhi, GU Lian-feng. Research Progress of Plant VIGS Technology and Its Application in Forestry Science[J]. Biotechnology Bulletin, 2021, 37(5): 141-153.
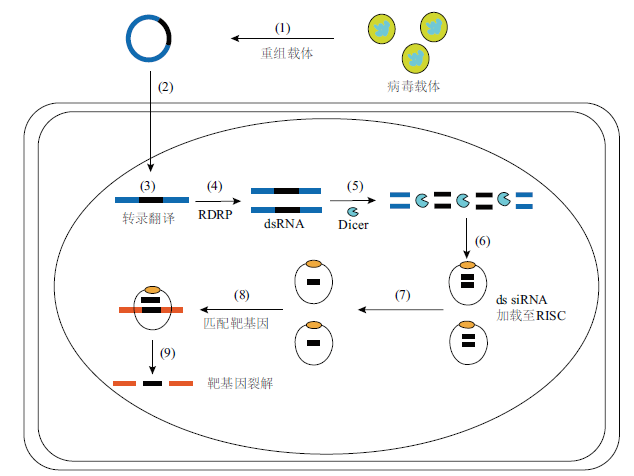
Fig.1 Molecular mechanism of VIGS (1) Construction of viral vectors. (2) Infection into plant cells. (3) Viral vectors are transcribed in the cytoplasm. (4) Synthesis of dsRNA by RDRP. (5)-(7) Dicer cut dsRNA into siRNA and then loaded it in RISC. (8)-(9) Activation of RISC target specific recognition of target genes resulted in host mRNA degradation and loss of gene expression
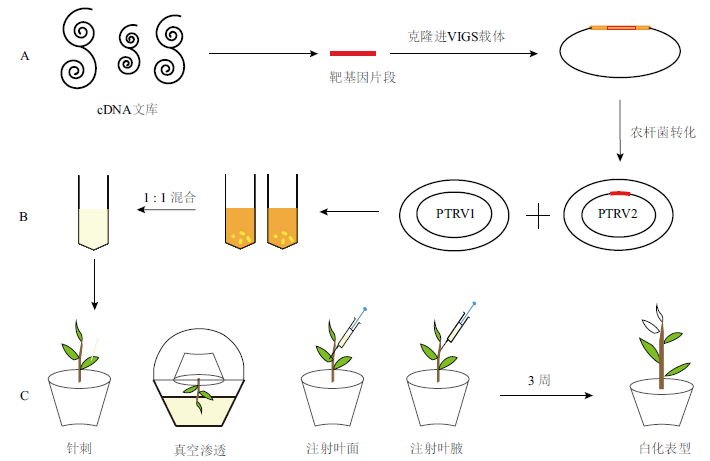
Fig. 2 Schematic diagram of VIGS infection technology A: The fragment of target gene (e.g. PDS) was cloned into the VIGS vector. B: Agrobacterium transformation culture PTRV1 and PTRV2 were mixed in equal proportion after resuspended. C: The photobleaching phenotype was observed after about 3 weeks after transformation into seedlings by means of toothpick, vacuum osmosis, or syringe injection to trigger plant defense mechanisms
| 软件名称 Software name | 软件介绍 Software introduction | 参考文献 Reference |
|---|---|---|
| SGN | 面向用户快速友好的交互式Web工具,在计算机中模拟细胞中发生的VIGS过程。帮助研究人员更快地设计VIGS构建载体,使用SGN工具可帮助选择沉默区域,识别最可能的目标和非目标位置,还可以进行预测沉默效率 | [ |
| pssRNAit | 利用全基因组数据设计特异性siRNA,可以进行全基因组脱靶评估是用于设计植物RNAi的有效的网络服务器工具。开发了针对RNAi途径的每个步骤的计算模型,有助设计沉默构建体 | [ |
| siRNA-Scan | 具有多个集成组件的搜索环境,包括序列相似性搜索,以预测载体序列是否会潜在脱靶和脱靶效率。可以搜索数据库中的其他脱靶基因然后选择特异性序列,用来设计具有最小脱靶概率的VIGS载体 | [ |
| DESIR | 设计siRNA的工具,基于siRNA反义链位置特异性核苷酸序列位置的线性模型进行设计。可得出siRNA设计结果以及预测沉默结果 http://biodev.extra.cea.fr/DSIR/DSIR.html | [ |
| DesiRm | 用于设计能够高效沉默基因的siRNA,可以提交mRNA以找出针对其的有效siRNA,以及提交siRNA及其靶序列以通过核苷酸错配设计更有效的siRNA | [ |
| i-Score | 对基因沉默RNA折叠剪切后所产生的siRNA进行效率检测,结果由高到低进行排序 | [ |
| DEQOR | 对基因区域能够发生较高沉默的区域进行预测,通过所选物种的转录组和基因组进行BLAST搜索,评估输入的靶基因位置的沉默效率 | [ |
| P-SAMS | web应用程序设计人工microRNA,有助于特异性的靶向不同的双子叶和单子叶植物物种中的一个以及多个基因 | [ |
| WMD3 | 基于web应用程序可用于设计人工microRNA构建基因沉默表达载体,可用目标搜索找到潜在的靶基因 | [ |
| PhieCBEs | 新型,高效,广靶向的植物编辑系统,是水稻和其他植物(作物)的单碱基编辑有效实用的工具,可在基因功能筛选,大规模饱和突变,编辑调控元件,RNA可变剪接,基因沉默等研究中广泛应用 | [ |
Table 1 Gene silence related design software
| 软件名称 Software name | 软件介绍 Software introduction | 参考文献 Reference |
|---|---|---|
| SGN | 面向用户快速友好的交互式Web工具,在计算机中模拟细胞中发生的VIGS过程。帮助研究人员更快地设计VIGS构建载体,使用SGN工具可帮助选择沉默区域,识别最可能的目标和非目标位置,还可以进行预测沉默效率 | [ |
| pssRNAit | 利用全基因组数据设计特异性siRNA,可以进行全基因组脱靶评估是用于设计植物RNAi的有效的网络服务器工具。开发了针对RNAi途径的每个步骤的计算模型,有助设计沉默构建体 | [ |
| siRNA-Scan | 具有多个集成组件的搜索环境,包括序列相似性搜索,以预测载体序列是否会潜在脱靶和脱靶效率。可以搜索数据库中的其他脱靶基因然后选择特异性序列,用来设计具有最小脱靶概率的VIGS载体 | [ |
| DESIR | 设计siRNA的工具,基于siRNA反义链位置特异性核苷酸序列位置的线性模型进行设计。可得出siRNA设计结果以及预测沉默结果 http://biodev.extra.cea.fr/DSIR/DSIR.html | [ |
| DesiRm | 用于设计能够高效沉默基因的siRNA,可以提交mRNA以找出针对其的有效siRNA,以及提交siRNA及其靶序列以通过核苷酸错配设计更有效的siRNA | [ |
| i-Score | 对基因沉默RNA折叠剪切后所产生的siRNA进行效率检测,结果由高到低进行排序 | [ |
| DEQOR | 对基因区域能够发生较高沉默的区域进行预测,通过所选物种的转录组和基因组进行BLAST搜索,评估输入的靶基因位置的沉默效率 | [ |
| P-SAMS | web应用程序设计人工microRNA,有助于特异性的靶向不同的双子叶和单子叶植物物种中的一个以及多个基因 | [ |
| WMD3 | 基于web应用程序可用于设计人工microRNA构建基因沉默表达载体,可用目标搜索找到潜在的靶基因 | [ |
| PhieCBEs | 新型,高效,广靶向的植物编辑系统,是水稻和其他植物(作物)的单碱基编辑有效实用的工具,可在基因功能筛选,大规模饱和突变,编辑调控元件,RNA可变剪接,基因沉默等研究中广泛应用 | [ |
| 树种Species | 病毒载体来源 Viral vector source | 参考文献 Reference |
|---|---|---|
| 胡杨(Populus euphratica) | TRV | [ |
| 银灰杨(Populus canescens) | TRV | [ |
| 麻风树(Jatropha curcas) | TRV | [ |
| 苹果(Malus domestica) | ALSV | [ |
| 木薯(Manihot esculenta) | ACMV | [ |
| 梨(Pyrus betulaefolia) 桃(Prunus persica) | ALSV,TRV TRV | [ [ |
| 樱桃(Prunus avium) | TRV | [ |
| 荔枝(Litchi chinensis) | TRV | [ |
| 欧洲越橘(Vaccinium myrtillus) | TRV | [ |
| 凤丹(Paeonia ostii) | TRV | [ |
| 橄榄(Oleaceae hoffmanns) | TRV | [ |
| 喜树(Camptotheca acuminata) | TRV | [ |
| 紫薇(Lagerstroemia indica) | TRV | [ |
Table 2 Research and application of VIGS system in poplar and other forest tree species
| 树种Species | 病毒载体来源 Viral vector source | 参考文献 Reference |
|---|---|---|
| 胡杨(Populus euphratica) | TRV | [ |
| 银灰杨(Populus canescens) | TRV | [ |
| 麻风树(Jatropha curcas) | TRV | [ |
| 苹果(Malus domestica) | ALSV | [ |
| 木薯(Manihot esculenta) | ACMV | [ |
| 梨(Pyrus betulaefolia) 桃(Prunus persica) | ALSV,TRV TRV | [ [ |
| 樱桃(Prunus avium) | TRV | [ |
| 荔枝(Litchi chinensis) | TRV | [ |
| 欧洲越橘(Vaccinium myrtillus) | TRV | [ |
| 凤丹(Paeonia ostii) | TRV | [ |
| 橄榄(Oleaceae hoffmanns) | TRV | [ |
| 喜树(Camptotheca acuminata) | TRV | [ |
| 紫薇(Lagerstroemia indica) | TRV | [ |
| [1] |
Zhao L, Zhang H, Kohnen MV, et al. Analysis of transcriptome and epitranscriptome in plants using pacbio iso-seq and nanopore-based direct RNA sequencing[J]. Frontiers in Genetics, 2019,10:253.
doi: 10.3389/fgene.2019.00253 URL |
| [2] |
Hofmann NR. Nanopore sequencing comes to plant genomes[J]. The Plant Cell, 2017,29(11):2677-2678.
doi: 10.1105/tpc.17.00863 pmid: 29114013 |
| [3] | Michael TP, Jackson SJTPG. The first 50 plant genomes[J]. The Plant Genome, 2013,6(2):6-13. |
| [4] |
Liu YJ, Wang XR, Zeng QY. De novo assembly of white poplar genome and genetic diversity of white poplar population in irtysh river basin in China[J]. Science China Life Sciences, 2019,62(5):609-618.
doi: 10.1007/s11427-018-9455-2 URL |
| [5] |
Nystedt B, Street NR, Wetterbom A, et al. The norway spruce genome sequence and conifer genome evolution[J]. Nature, 2013,497(7451):579-584.
doi: 10.1038/nature12211 pmid: 23698360 |
| [6] |
Ye G, Zhang H, Chen B, et al. De novo genome assembly of the stress tolerant forest species casuarina equisetifolia provides insight into secondary growth[J]. The Plant Journal, 2019,97(4):779-794.
doi: 10.1111/tpj.2019.97.issue-4 URL |
| [7] |
Busov VB, Brunner AM, Meilan R, et al. Genetic transformation:a powerful tool for dissection of adaptive traits in trees[J]. New Phytol, 2005,167(1):9-18.
doi: 10.1111/j.1469-8137.2005.01412.x URL |
| [8] |
Ye S, Cai C, Ren H, et al. An efficient plant regeneration and transformation system of ma bamboo(Dendrocalamus latiflorus munro)started from young shoot as explant[J]. Frontiers in Plant Science, 2017,8:1298.
doi: 10.3389/fpls.2017.01298 URL |
| [9] | 瞿礼嘉, 王小菁, 王台, 等. 2007 年中国植物科学若干领域重要研究进展[J]. 植物学报, 2009,44(1):2-26. |
| Qu LJ, Wang XJ, Wang T, et al. Research advances on plant science in china in 2007[J]. Chinese Bulletin of Botany, 2009,44(1):2-26 | |
| [10] | Pandey P, Senthil M, Mysore KS. Advances in plant gene silencing methods[J]. Methods Mol Biol, 2015,1287:3-23. |
| [11] |
Baulcombe DC. Fast forward genetics based on virus-induced gene silencing[J]. Curr Opin Plant Biol, 1999,2(2):109-113.
pmid: 10322199 |
| [12] |
Cui H, Wang A. An efficient viral vector for functional genomic studies of prunus fruit trees and its induced resistance to plum pox virus via silencing of a host factor gene[J]. Plant Biotechnol J, 2017,15(3):344-356.
doi: 10.1111/pbi.2017.15.issue-3 URL |
| [13] |
Anandalakshmi R, Pruss GJ, Ge X, et al. A viral suppressor of gene silencing in plants[J]. Proc Natl Acad Sci USA, 1998,95(22):13079-13084.
doi: 10.1073/pnas.95.22.13079 URL |
| [14] |
Vance V, Vaucheret H. RNA silencing in plants defense and counterdefense[J]. Science, 2001,292(5525):2277-2280.
doi: 10.1126/science.1061334 URL |
| [15] |
Kumagai MH, Donson J, Della G, et al. Cytoplasmic inhibition of carotenoid biosynjournal with virus-derived rna[J]. Proc Natl Acad Sci USA, 1995,92(5):1679-1683.
doi: 10.1073/pnas.92.5.1679 URL |
| [16] |
Szittya G, Burgyán J. RNA interference-mediated intrinsic antiviral immunity in plants[J]. Current Topics in Microbiology and Immunology, 2013,371:153-181.
doi: 10.1007/978-3-642-37765-5_6 pmid: 23686235 |
| [17] |
Oliva R, Win J, Raffaele S, et al. Recent developments in effector biology of filamentous plant pathogens[J]. Cell Microbiol, 2010,12(6):705-715.
doi: 10.1111/cmi.2010.12.issue-6 URL |
| [18] |
Yamagishi N, Yoshikawa N. Virus-induced gene silencing as a tool for analysis of gene functions in plants[J]. Uirusu, 2010,60(2):155-162.
doi: 10.2222/jsv.60.155 URL |
| [19] |
Qin C, Li B, Fan Y, et al. Roles of dicer-like proteins 2 and 4 in intra-and intercellular antiviral silencing[J]. Plant Physiol, 2017,174(2):1067-1081.
doi: 10.1104/pp.17.00475 URL |
| [20] | Kobayashi H, Tomari Y. Risc assembly:coordination between small rnas and argonaute proteins[J]. Biochimica et Biophysica Acta, 2016,1859(1):71-81. |
| [21] |
Dommes AB, Gross T, Herbert DB, et al. Virus-induced gene silencing:empowering genetics in non-model organisms[J]. J Exp Bot, 2019,70(3):757-770.
doi: 10.1093/jxb/ery411 |
| [22] |
Burch TM, Anderson JC, Martin GB, et al. Applications and advantages of virus-induced gene silencing for gene function studies in plants[J]. Plant J, 2004,39(5):734-746.
doi: 10.1111/tpj.2004.39.issue-5 URL |
| [23] |
Liu Y, Schiff M, Dinesh SP. Virus-induced gene silencing in tomato[J]. Plant J, 2002,31(6):777-786.
doi: 10.1046/j.1365-313X.2002.01394.x URL |
| [24] |
Singh DK, Lee HK, Dweikat I, et al. An efficient and improved method for virus-induced gene silencing in sorghum[J]. BMC Plant Biol, 2018,18(1):123.
doi: 10.1186/s12870-018-1344-z URL |
| [25] |
Liu E, Page JE. Optimized cDNA libraries for virus-induced gene silencing(vigs)using tobacco rattle virus[J]. Plant Methods, 2008,4:5.
doi: 10.1186/1746-4811-4-5 URL |
| [26] |
Peele C, Jordan CV, Muangsan N, et al. Silencing of a meristematic gene using geminivirus-derived vectors[J]. Plant J, 2001,27(4):357-366.
pmid: 11532181 |
| [27] |
Turnage MA, Muangsan N, Peele CG, et al. Geminivirus-based vectors for gene silencing in Arabidopsis[J]. Plant J, 2002,30(1):107-114.
doi: 10.1046/j.1365-313X.2002.01261.x URL |
| [28] | Dinesh SP, Anandalakshmi R, Marathe R, et al. Virus-induced gene silencing[J]. Methods Mol Biol, 2003,236:287-294. |
| [29] |
Lentz EM, Kuon JE, Alder A, et al. Cassava geminivirus agroclones for virus-induced gene silencing in cassava leaves and roots[J]. Plant Methods, 2018,14:73.
doi: 10.1186/s13007-018-0340-5 URL |
| [30] |
Rosa C, Kuo YW, Wuriyanghan H, et al. RNA interference mechanisms and applications in plant pathology[J]. Annual Review of Phytopathology, 2018,56:581-610.
doi: 10.1146/annurev-phyto-080417-050044 URL |
| [31] |
Yang J, Zhang TY, Liao QS, et al. Chinese wheat mosaic virus-induced gene silencing in monocots and dicots at low temperature[J]. Front Plant Sci, 2018,9:1627.
doi: 10.3389/fpls.2018.01627 URL |
| [32] |
Biruma M, Martin T, Fridborg I, et al. Two loci in sorghum with NB-LRR encoding genes confer resistance to Colletotrichum sublineolum[J]. Theoretical and Applied Genetics, 2012,124(6):1005-1015.
doi: 10.1007/s00122-011-1764-8 URL |
| [33] |
Martin T, Biruma M, Fridborg I, et al. A highly conserved NB-LRR encoding gene cluster effective against setosphaeria turcica in sorghum[J]. BMC Plant Biol, 2011,11:151.
doi: 10.1186/1471-2229-11-151 pmid: 22050783 |
| [34] |
Malamy J, Hennig J, Klessig DF. Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection[J]. The Plant Cell, 1992,4(3):359-366.
pmid: 12297650 |
| [35] | Velásquez AC, Chakravarthy S, Martin GB. Virus-induced gene silencing(vigs)in Nicotiana benthamiana and tomato[J]. Journal of Visualized Experiments, 2009(28):1292. |
| [36] | Fu DQ, Zhu B, Zhu H, et al. Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity[J]. Mol Cells, 2006,21(1):153-160. |
| [37] |
Baron C, Domke N, Beinhofer M, et al. Elevated temperature differentially affects virulence, virb protein accumulation, and t-pilus formation in different agrobacterium tumefaciens and agrobacterium vitis strains[J]. Journal of Bacteriology, 2001,183(23):6852-6861.
doi: 10.1128/JB.183.23.6852-6861.2001 URL |
| [38] |
Pacak A, Strozycki PM, Barciszewska M, et al. The brome mosaic virus-based recombination vector triggers a limited gene silencing response depending on the orientation of the inserted sequence[J]. Archives of Virology, 2010,155(2):169-179.
doi: 10.1007/s00705-009-0556-9 URL |
| [39] |
Xu H, Xu L, Yang P, et al. Tobacco rattle virus-induced PDS and ChlH gene silencing in solanum[J]. PeerJ, 2018,6:e4424.
doi: 10.7717/peerj.4424 URL |
| [40] | Saitoh H, Terauchi R. Virus-induced silencing of Ftsh gene in Nicotiana benthmiana causes a striking bleached leaf phenotype[J]. Genes & Genetic Systems, 2002,77(5):335-340. |
| [41] |
Kjemtrup S, Sampson KS, Peele CG, et al. Gene silencing from plant DNA carried by a geminivirus[J]. Plant J, 1998,14(1):91-100.
pmid: 15494056 |
| [42] |
Liu H, Fu D, Zhu B, et al. Virus-induced gene silencing in eggplant(Solanum melongena)[J]. Journal of Integrative Plant Biology, 2012,54(6):422-429.
doi: 10.1111/jipb.2012.54.issue-6 URL |
| [43] |
Rotenberg D, Thompson TS, German TL, et al. Methods for effective real-time rt-pcr analysis of virus-induced gene silencing[J]. J Virol Methods, 2006,138(1-2):49-59.
pmid: 16959330 |
| [44] |
Fernandez-Pozo N, Rosli HG, Martin GB, et al. The sgn vigs tool:user-friendly software to design virus-induced gene silencing(vigs)constructs for functional genomics[J]. Mol Plant, 2015,8(3):486-488.
doi: 10.1016/j.molp.2014.11.024 pmid: 25667001 |
| [45] |
Ahmed F, Senthil M, Dai X, et al. Pssrnait:a web server for designing effective and specific plant sirnas with genome-wide off-target assessment[J]. Plant Physiol, 2020,184(1):65-81.
doi: 10.1104/pp.20.00293 URL |
| [46] |
Xu P, Zhang Y, Kang L, et al. Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants[J]. Plant Physiol, 2006,142(2):429-440.
doi: 10.1104/pp.106.083295 URL |
| [47] |
Vert JP, Foveau N, Lajaunie C, et al. An accurate and interpretable model for sirna efficacy prediction[J]. BMC Bioinformatics, 2006,7:520.
doi: 10.1186/1471-2105-7-520 URL |
| [48] |
Ahmed F, Raghava GP. Designing of highly effective complementary and mismatch sirnas for silencing a gene[J]. PLoS One, 2011,6(8):e23443.
doi: 10.1371/journal.pone.0023443 URL |
| [49] |
Ichihara M, Murakumo Y, Masuda A, et al. Thermodynamic instability of sirna duplex is a prerequisite for dependable prediction of sirna activities[J]. Nucleic Acids Res, 2007,35(18):e123.
doi: 10.1093/nar/gkm699 URL |
| [50] |
Henschel A, Buchholz F, Habermann B. Deqor:a web-based tool for the design and quality control of sirnas[J]. Nucleic Acids Res, 2004,32(Web Server issue):W113-W120.
doi: 10.1093/nar/gkh408 URL |
| [51] | Fahlgren N, Hill ST, Carrington JC, et al. P-sams:a web site for plant artificial microrna and synthetic trans-acting small interfering rna design[J]. Bioinformatics(Oxford, England), 2016,32(1):157-158. |
| [52] |
Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial micrornas and other small rnas[J]. Plant J, 2008,53(4):674-690.
doi: 10.1111/j.1365-313X.2007.03328.x URL |
| [53] |
Zeng D, Liu T, Tan J, et al. Phiecbes:plant high-efficiency cytidine base editors with expanded target range[J]. Mol Plant, 2020,13(12):1666-1669.
doi: 10.1016/j.molp.2020.11.001 URL |
| [54] |
Tuskan GA, Difazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa[J]. Science, 2006,313(5793):1596-1604.
doi: 10.1126/science.1128691 URL |
| [55] |
Jansson S, Douglas CJ. Populus:a model system for plant biology[J]. Annual Review of Plant Biology, 2007,58:435-458.
pmid: 17280524 |
| [56] |
Shen Z, Sun J, Yao J, et al. High rates of virus-induced gene silencing by tobacco rattle virus in populus[J]. Tree Physiology, 2015,35(9):1016-1029.
doi: 10.1093/treephys/tpv064 URL |
| [57] |
Ye J, Qu J, Bui HTN, et al. Rapid analysis of Jatropha curcas gene functions by virus-induced gene silencing[J]. Plant Biotechnol J, 2009,7(9):964-976.
doi: 10.1111/pbi.2009.7.issue-9 URL |
| [58] |
Sasaki S, Yamagishi N, Yoshikawa N. Efficient virus-induced gene silencing in apple, pear and japanese pear using apple latent spherical virus vectors[J]. Plant Methods, 2011,7(1):15.
doi: 10.1186/1746-4811-7-15 URL |
| [59] |
Fofana IB, Sangaré A, Collier R, et al. A geminivirus-induced gene silencing system for gene function validation in cassava[J]. Plant Mol Biol, 2004,56(4):613-624.
doi: 10.1007/s11103-004-0161-y URL |
| [60] |
Hao L, Zhang Y, Wang S, et al. A constitutive and drought-responsive mrna undergoes long-distance transport in pear(Pyrus betulaefolia)phloem[J]. Plant Science, 2020,293:110419.
doi: 10.1016/j.plantsci.2020.110419 URL |
| [61] | Jia H, Guo J, Qin L, et al. Virus-induced PpCHLH gene silencing in peach leaves(Prunus persica)[J]. Journal of Pomology & Horticultural Science, 2010,85(6):528-532. |
| [62] |
Shen X, Zhao K, Liu L, et al. A role for pacmyba in aba-regulated anthocyanin biosynjournal in red-colored sweet cherry cv. Hong deng(Prunus avium L.)[J]. Plant Cell Physiol, 2014,55(5):862-880.
doi: 10.1093/pcp/pcu013 URL |
| [63] |
Wang C, Lü P, Zhong S, et al. Lcmcii-1 is involved in the ros-dependent senescence of the rudimentary leaves of Litchi chinensis[J]. Plant Cell Reports, 2017,36(1):89-102.
doi: 10.1007/s00299-016-2059-y URL |
| [64] |
Jaakola L, Poole M, Jones MO, et al. A squamosa mads box gene involved in the regulation of anthocyanin accumulation in bilberry fruits[J]. Plant Physiol, 2010,153(4):1619-1629.
doi: 10.1104/pp.110.158279 URL |
| [65] |
Xie L, Zhang Q, Sun D, et al. Virus-induced gene silencing in the perennial woody[J]. PeerJ, 2019,7:e7001.
doi: 10.7717/peerj.7001 URL |
| [66] | Koudounas K, Thomopoulou M, Angeli E, et al. Virus-induced gene silencing in olive tree(Oleaceae)[J]. Methods Mol Biol, 2020,2172:165-182. |
| [67] |
Jin Z, Cong Y, Zhu S, et al. Two classes of cytochrome p450 reductase genes and their divergent functions in Camptotheca acuminata decne[J]. International Journal of Biological Macromolecules, 2019,138:1098-1108.
doi: 10.1016/j.ijbiomac.2019.07.141 URL |
| [68] |
Li S, Zheng T, Zhuo X, et al. Transcriptome profiles Reveal that gibberellin-related genes regulate weeping traits in crape myrtle[J]. Horticulture Research, 2020,7:54.
doi: 10.1038/s41438-020-0279-3 URL |
| [69] |
Ratcliff F, Martin-Hernandez AM, Baulcombe DC. Technical advance. Tobacco rattle virus as a vector for analysis of gene function by silencing[J]. Plant J, 2001,25(2):237-245.
pmid: 11169199 |
| [70] | Li C, Yamagishi N, Yoshikawa N. Rna silencing-mediated apple latent spherical virus vaccine in plants[J]. Methods Mol Biol, 2019,2028:273-288. |
| [71] | Li C, Yoshikawa N, Takahashi T, et al. Nucleotide sequence and genome organization of apple latent spherical virus:a new virus classified into the family comoviridae[J]. The Journal of General Virology, 2000,81(Pt 2):541-547. |
| [72] |
Igarashi A, Yamagata K, Sugai T, et al. Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes[J]. Virology, 2009,386(2):407-416.
doi: 10.1016/j.virol.2009.01.039 URL |
| [73] |
Yamagishi N, Yoshikawa N. Virus-induced gene silencing in soybean seeds and the emergence stage of soybean plants with apple latent spherical virus vectors[J]. Plant Mol Biol, 2009,71(1-2):15-24.
doi: 10.1007/s11103-009-9505-y URL |
| [74] | Yamagishi N, Yoshikawa N. Highly efficient virus-induced gene silencing in apple and soybean by apple latent spherical virus vector and biolistic inoculation[J]. Methods Mol Biol, 2013,975:167-181. |
| [75] |
Ceballos H, Iglesias CA, Pérez JC, et al. Cassava breeding:opportunities and challenges[J]. Plant Mol Biol, 2004,56(4):503-516.
pmid: 15630615 |
| [76] |
Zhang J, Xie M, Tuskan GA, et al. Recent advances in the transcriptional regulation of secondary cell wall biosynjournal in the woody plants[J]. Front Plant Sci, 2018,9:1535.
doi: 10.3389/fpls.2018.01535 pmid: 30405670 |
| [77] |
Pandey SK, Nookaraju A, Fujino T, et al. Virus-induced gene silencing(vigs)-mediated functional characterization of two genes involved in lignocellulosic secondary cell wall formation[J]. Plant Cell Reports, 2016,35(11):2353-2367.
doi: 10.1007/s00299-016-2039-2 URL |
| [78] |
Liu J, Zeng Y, Yan P, et al. Transcriptional and hormonal regulation of weeping trait in salix matsudana[J]. Genes, 2017,8(12):359.
doi: 10.3390/genes8120359 URL |
| [79] |
Hakoshima T. Structural basis of the specific interactions of gras family proteins[J]. FEBS Letters, 2018,592(4):489-501.
doi: 10.1002/1873-3468.12987 pmid: 29364510 |
| [80] |
Burch TM, Schiff M, Liu Y, et al. Efficient virus-induced gene silencing in Arabidopsis[J]. Plant Physiol, 2006,142(1):21-27.
doi: 10.1104/pp.106.084624 URL |
| [81] |
Zhou B, Zeng L. Elucidating the role of highly homologous ubiquitin E2 gene family members in plant immunity through an improved virus-induced gene silencing approach[J]. Plant Methods, 2017,13:59.
doi: 10.1186/s13007-017-0210-6 URL |
| [82] | Chen W, Zhang Q, Kong J, et al. Mr vigs:Microrna-based virus-induced gene silencing in plants[J]. Methods Mol Biol, 2015,1287:147-157. |
| [83] |
Wu C, Jia L, Goggin F. The reliability of virus-induced gene silencing experiments using tobacco rattle virus in tomato is influenced by the size of the vector control[J]. Mol Plant Pathol, 2011,12(3):299-305.
doi: 10.1111/mpp.2011.12.issue-3 URL |
| [84] |
Cakir C, Tör M. Factors influencing barley stripe mosaic virus-mediated gene silencing in wheat[J]. Physiological and Molecular Plant Pathology, 2010,74(3-4):246-253.
doi: 10.1016/j.pmpp.2010.04.001 URL |
| [85] |
Bruun M, Madsen CT, Jessing S, et al. Stability of barley stripe mosaic virus-induced gene silencing in barley[J]. Mol Plant Microbe Interact, 2007,20(11):1323-1331.
doi: 10.1094/MPMI-20-11-1323 URL |
| [86] |
Senthil M, Mysore KS. New dimensions for vigs in plant functional genomics[J]. Trends Plant Sci, 2011,16(12):656-665.
doi: 10.1016/j.tplants.2011.08.006 URL |
| [87] | Senthil M. Virus-induced gene silencing and its applications[J]. CAB Reviews:Perspectives in Agriculture, Veterinary Science, Nutrition and Natural Resources, 2008,3(11):1-18. |
| [88] |
Senthil M, Mysore KS. Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato[J]. Plant Biotechnol J, 2011,9(7):797-806.
doi: 10.1111/pbi.2011.9.issue-7 URL |
| [89] |
Yamagishi N, Yoshikawa N. Virus-induced gene silencing in soybean seeds and the emergence stage of soybean plants with apple latent spherical virus vectors[J]. Plant Mol Biol, 2009,71(1-2):15-24.
doi: 10.1007/s11103-009-9505-y URL |
| [90] |
Ali MS, Baek KH. Co-suppression of NbClpC1 and NbClpC2 alters plant morphology with changed hormone levels in Nicotiana benthamiana[J]. Plant Cell Reports, 2019,38(10):1317-1328.
doi: 10.1007/s00299-019-02452-8 URL |
| [91] | Gunupuru LR, Perochon A, Ali SS, et al. Virus-induced gene silencing(vigs)for functional characterization of disease resistance genes in barley seedlings[J]. Methods Mol Biol, 2019,1900:95-116. |
| [92] |
Sui X, Zhao M, Han X, et al. Rrgt1, a key gene associated with anthocyanin biosynjournal, was isolated from rosa rugosa and identified via overexpression and vigs[J]. Plant Physiology and Biochemistry, 2019,135:19-29.
doi: 10.1016/j.plaphy.2018.11.022 URL |
| [93] |
Ahn CS, Lee JH, Reum Hwang A, et al. Prohibitin is involved in mitochondrial biogenesis in plants[J]. Plant J, 2006,46(4):658-667.
doi: 10.1111/tpj.2006.46.issue-4 URL |
| [94] |
Liu Y, Nakayama N, Schiff M, et al. Virus induced gene silencing of a deficiens ortholog in Nicotiana benthamiana[J]. Plant Mol Biol, 2004,54(5):701-711.
doi: 10.1023/B:PLAN.0000040899.53378.83 URL |
| [95] | Jiang J, Ma S, Ye N, et al. Wrky transcription factors in plant responses to stresses[J]. Journal of Integrative Plant Biology, 2017,59(2). |
| [96] |
Ekengren SK, Liu Y, Schiff M, et al. Two mapk cascades, npr1, and tga transcription factors play a role in pto-mediated disease resistance in tomato[J]. Plant J, 2003,36(6):905-917.
pmid: 14675454 |
| [97] |
He X, Anderson JC, Del O, et al. Silencing of subfamily i of protein phosphatase 2a catalytic subunits results in activation of plant defense responses and localized cell death[J]. Plant J, 2004,38(4):563-577.
doi: 10.1111/tpj.2004.38.issue-4 URL |
| [98] |
Mali P, Yang L, Esvelt KM, et al. Rna-guided human genome engineering via cas9[J]. Science, 2013,339(6121):823-826.
doi: 10.1126/science.1232033 URL |
| [99] |
Ali Z, Abul A, Li L, et al. Efficient virus-mediated genome editing in plants using the crispr/cas9 system[J]. Mol Plant, 2015,8(8):1288-1291.
doi: 10.1016/j.molp.2015.02.011 URL |
| [100] |
Yin K, Han T, Liu G, et al. A geminivirus-based guide rna delivery system for crispr/cas9 mediated plant genome editing[J]. Scientific Reports, 2015,5:14926.
doi: 10.1038/srep14926 URL |
| [101] |
Ellison EE, Nagalakshmi U, Gamo ME, et al. Multiplexed heritable gene editing using rna viruses and mobile single guide rnas[J]. Nat Plants, 2020,6(6):620-624.
doi: 10.1038/s41477-020-0670-y pmid: 32483329 |
| [102] | Jackson SD, Hong Y. Systemic movement of ft mrna and a possible role in floral induction[J]. Front Plant Sci, 2012,3:127. |
| [103] |
Christiaens O, Whyard S, Vélez AM, et al. Double-stranded rna technology to control insect pests:current status and challenges[J]. Front Plant Sci, 2020,11:451.
doi: 10.3389/fpls.2020.00451 pmid: 32373146 |
| [1] | LIU Zhen-yin, DUAN Zhi-zhen, PENG Ting, WANG Tong-xin, WANG Jian. Establishment and Optimization of Virus-induced Gene Silencing System in Bougainvillea peruviana ‘Thimma’ [J]. Biotechnology Bulletin, 2023, 39(7): 123-130. |
| [2] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [3] | WU Qin-qin, SUN Min, CHEN Yu, FU Ya-qin, ZENG Bin, HE Bin. Strategies and Advances in Functional Genomics of Aspergillus oryzae [J]. Biotechnology Bulletin, 2019, 35(8): 186-192. |
| [4] | LIU Guo-hong, LIU Qin-ying, LIU Bo, CHE Jian-mei, CHEN Qian-qian. Investigation of Bacillus Resources in the Soil of Agroforestry Biome:Case Study in Some Parts of Sichuan and Chongqing [J]. Biotechnology Bulletin, 2019, 35(3): 78-86. |
| [5] | LEI Zhao-xia, LIU Jing, BAI Yi-ping, TANG Wei, WANG Hong-yang. Cloning and Functional Analysis of a Potato Ubiquitin-conjugating Enzyme Gene StUBC17 [J]. Biotechnology Bulletin, 2019, 35(1): 35-41. |
| [6] | Sun Wei, Xu Yi, Xu Guiying, Sun Peiguang, Song Shun, Chang Shenghe. Virus-induced Gene Silencing and Its Application in Plant Research [J]. Biotechnology Bulletin, 2015, 31(10): 105-110. |
| [7] | Zhang Zhaojun, Wang Xiaobin, Wang Hui, Liu Lin, Zhang Xinge, He Xiuxia. Study on Chinese Tomato Yellow Leaf Curl Virus-induced Gene Silencing(VIGS)via Root Absorption [J]. Biotechnology Bulletin, 2014, 0(1): 143-146. |
| [8] | Xing Guofang, Guo Pingyi. Functional Genomics Approaches for the Study of Low Phosphate Responses in Higher Plants [J]. Biotechnology Bulletin, 2013, 0(7): 1-6. |
| [9] | Zhang Yanming, Xing Guofang, Liu Meitao, Liu Xiaodong, Han Yuanhuai. Genome Wide Association Study: Opportunities and Challenges in Genomic Research [J]. Biotechnology Bulletin, 2013, 0(6): 1-6. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||