








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (10): 120-127.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1497
Previous Articles Next Articles
XIONG Liang-bin1,2( ), SUN Ji1, LIU Xian-zhou1, QU Zhan-guo1, JI Yu-qing1, XU Yi-xin1(
), SUN Ji1, LIU Xian-zhou1, QU Zhan-guo1, JI Yu-qing1, XU Yi-xin1( ), WANG Feng-qing2(
), WANG Feng-qing2( )
)
Received:2020-12-09
Online:2021-10-26
Published:2021-11-12
Contact:
XU Yi-xin,WANG Feng-qing
E-mail:lbxiong2010@163.com;xuyx@sumhs.edu.cn;fqwang@ecust.edu.cn
XIONG Liang-bin, SUN Ji, LIU Xian-zhou, QU Zhan-guo, JI Yu-qing, XU Yi-xin, WANG Feng-qing. Transcription Variations of Key Genes in the Central Metabolism Pathways of the Sterols Transformation in Mycobacterium[J]. Biotechnology Bulletin, 2021, 37(10): 120-127.
| 名称Name | 描述Description | 来源Source |
|---|---|---|
| 菌株 | ||
| Escherichia coli DH5α | 用于质粒转化扩增 | 天根生化 |
| Mycobacterium neoaurum ATCC 25795(Mn) | 野生型菌株 | ATCC |
| MnΔkstD1(MnΔk1) | 在野生型Mn菌株中删除kstD1基因,具有转化甾醇积累9-OHAD的生产能力 | 姚抗等[ |
| 质粒 | ||
| p2NIL | 用于分枝杆菌基因敲除的同源臂构建载体,KanR | Dr. Parish惠赠 |
| p2N-kstD1 | 携带kstD1基因敲除同源臂的p2NIL重组质粒 | 姚抗等[ |
| pGOAL19 | HygR, Pag85-lacZ, Phsp60-sacB, Pac I cassette vector, AmpR | Dr. Parish惠赠 |
| p19-kstD1 | p2NIL携带kstD1基因敲除同源臂及pGOAL19筛选表达框组成的重组质粒,KanR和HygR | 姚抗等[ |
Table 1 Strains and plasmids used in this study
| 名称Name | 描述Description | 来源Source |
|---|---|---|
| 菌株 | ||
| Escherichia coli DH5α | 用于质粒转化扩增 | 天根生化 |
| Mycobacterium neoaurum ATCC 25795(Mn) | 野生型菌株 | ATCC |
| MnΔkstD1(MnΔk1) | 在野生型Mn菌株中删除kstD1基因,具有转化甾醇积累9-OHAD的生产能力 | 姚抗等[ |
| 质粒 | ||
| p2NIL | 用于分枝杆菌基因敲除的同源臂构建载体,KanR | Dr. Parish惠赠 |
| p2N-kstD1 | 携带kstD1基因敲除同源臂的p2NIL重组质粒 | 姚抗等[ |
| pGOAL19 | HygR, Pag85-lacZ, Phsp60-sacB, Pac I cassette vector, AmpR | Dr. Parish惠赠 |
| p19-kstD1 | p2NIL携带kstD1基因敲除同源臂及pGOAL19筛选表达框组成的重组质粒,KanR和HygR | 姚抗等[ |
| 用途 Purpose | 名称Name | 描述Description |
|---|---|---|
| 基因删除引物 Primers for gene deletion | D-kstD1-UF-Hind III | CAGTaagcttCTTCTCAGCCATACGTGGCTCCTA |
| D-kstD1-UR-Pst I | ttaactgcaggtcctgggcagtcatgtagaacac | |
| D-kstD1-DF-Pst I | tacactgcagttgcatctcgctggaaaggcctga | |
| D-kstD1-DR-Kpn I | TATAggtaccCGCGGTCAGCGTTCCGATGAACTT | |
| qRT-PCR分析引物 Primers for qRT-PCR analysis | R-16S rRNA-F | CCTATGTTGCCAGCGGGTTATGC & GCGATTACTAGCGACTCCGACTTCA |
| R-pfkA-F & R | CCAGGCTTGAACGCCGTGATCAG & GATCGGGATCAACACATCGATGC | |
| R-pfkB-F & R | TACCTGCTGCTGCCGACGATCCG & ACCACGTCGGCGTACCAGCTCGT | |
| R-pyk-F & R | GACCACGAAGAGTCCTATCGCCG & ACCAGTCCGACGTTGCCGTCGTC | |
| R-zwf-F & R | CTGATGCCGGCGATCTACGACCT & GTAGAACGCGTGATTGCCGTTGG | |
| R-gntZ-F & R | CTGACCGAAGGCTACGGATTGAT & GCAGCCAGCATCTCGTAACCCTC | |
| R-citA-F & R | GACATCGACGACCTGGTATCGGA & ACATAGGACAGCGCCATCGCCGA | |
| R-icd2-F & R | ATCAAGCTGCCGAACATCAGCGC & TGGCCCATGCTGTGCGGGTGCTT | |
| R-kdg-F & R | CGGCCAAGGCGATGATCGACAAC & CATCGTCTCGCACTTCTTGATGG |
Table 2 Primers used in this study
| 用途 Purpose | 名称Name | 描述Description |
|---|---|---|
| 基因删除引物 Primers for gene deletion | D-kstD1-UF-Hind III | CAGTaagcttCTTCTCAGCCATACGTGGCTCCTA |
| D-kstD1-UR-Pst I | ttaactgcaggtcctgggcagtcatgtagaacac | |
| D-kstD1-DF-Pst I | tacactgcagttgcatctcgctggaaaggcctga | |
| D-kstD1-DR-Kpn I | TATAggtaccCGCGGTCAGCGTTCCGATGAACTT | |
| qRT-PCR分析引物 Primers for qRT-PCR analysis | R-16S rRNA-F | CCTATGTTGCCAGCGGGTTATGC & GCGATTACTAGCGACTCCGACTTCA |
| R-pfkA-F & R | CCAGGCTTGAACGCCGTGATCAG & GATCGGGATCAACACATCGATGC | |
| R-pfkB-F & R | TACCTGCTGCTGCCGACGATCCG & ACCACGTCGGCGTACCAGCTCGT | |
| R-pyk-F & R | GACCACGAAGAGTCCTATCGCCG & ACCAGTCCGACGTTGCCGTCGTC | |
| R-zwf-F & R | CTGATGCCGGCGATCTACGACCT & GTAGAACGCGTGATTGCCGTTGG | |
| R-gntZ-F & R | CTGACCGAAGGCTACGGATTGAT & GCAGCCAGCATCTCGTAACCCTC | |
| R-citA-F & R | GACATCGACGACCTGGTATCGGA & ACATAGGACAGCGCCATCGCCGA | |
| R-icd2-F & R | ATCAAGCTGCCGAACATCAGCGC & TGGCCCATGCTGTGCGGGTGCTT | |
| R-kdg-F & R | CGGCCAAGGCGATGATCGACAAC & CATCGTCTCGCACTTCTTGATGG |
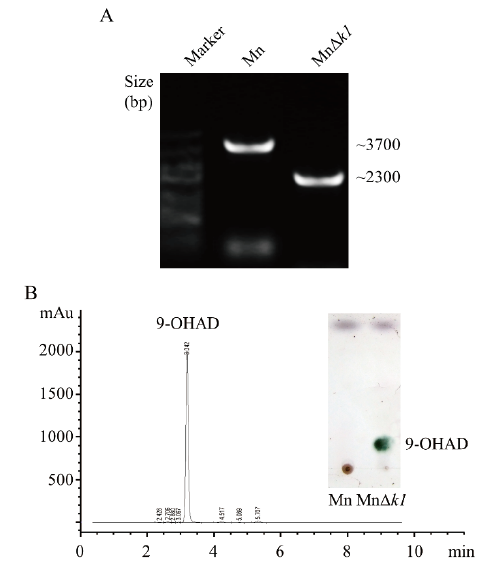
Fig. 1 Effect of deleting kstD1 on the Mycobacterium PCR verification and biotransformation of sterols A: Evidence for allelic replacement of kstD1 gene. Mn: The wild-type strain. MnΔk1: Deletion of kstD1 in the Mn strain. B: HPLC and TLC analysis results of the sterol biotransformation in MnΔk1 strain
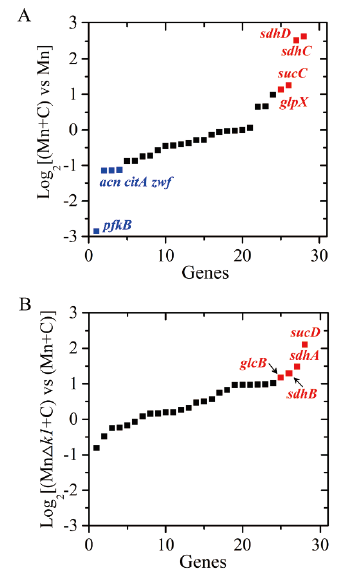
Fig. 2 Transcriptional comparisons of genes related in central metabolism pathways during the sterol transformation in Mycobacterium A: Transcript profiles of the genes in wild-type Mn strain. B: Transcript profiles of the genes in MnΔk1 strain. Mn: The wild type strain was cultivated in fermentation medium without phytosterol addition. Mn+C: The wild type strain was cultured in fermentation medium with 1 g/L of phytosterol addition. MnΔk1+C: the strain MnΔk1 was cultured in fermentation medium with 1 g/L of phytosterol addition
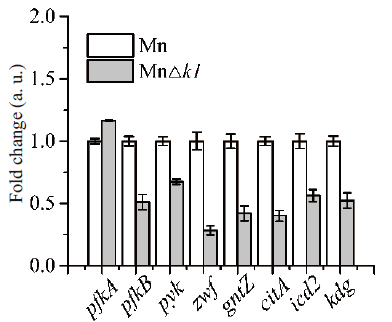
Fig. 3 qRT-PCR analysis of key genes in central metabolism pfkA & pfkB: 6-phosphofructokinase; pyk: pyruvate kinase; zwf: glucose 6-phosphatedehydrogenase; gntZ: 6-phosphogluconate dehydrogenase; citA: citrate synthase; icd2: isocitric dehydrogenase; kdg: oxoglutarate dehydrogenase
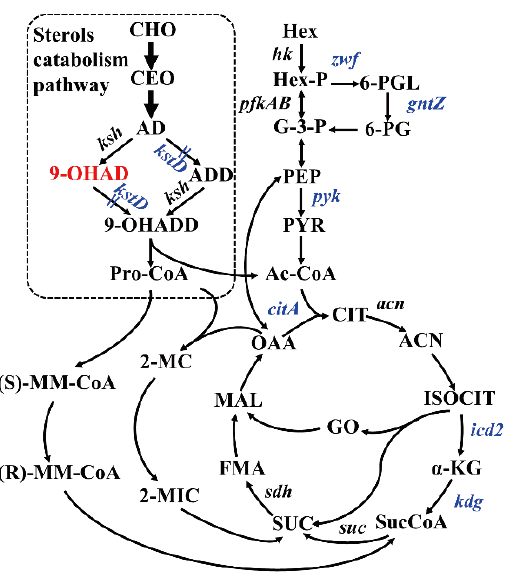
Fig. 5 Schematic diagram of sterol metabolism pathway and central metabolism pathway in Mycobacterium CHO: cholesterol; CEO: cholesterone; AD: 4-androstene-3,17-dione; ADD: 1,4-androstadiene-3,17-dione; 9-OHAD: 9α-hydroxy-4-androstene-3,17-dione; 9-OHADD: 9 α-hydroxy-1,4-androstadiene-3,17-dione; Pro-CoA: propionyl-CoA; Hex: hexose; Hex-P: hexose phosphate; G-3-P: glyceraldehyde 3-phosphate; PEP: phosphoenolpyruvate; PYR: pyruvic acid; Ac-CoA: acetyl-CoA; (S)-MM-CoA: (S)-methylmalonyl-CoA; (R)-MM-CoA: (R)-methylmalonyl-CoA; 2-MC: 2-methycitrate; 2-MIC: 2-methylisocitrate; CIT: citrate; ACN: aconitic acid; ISOCIT: isocitrate; α-KG: α-ketoglutarate; SucCoA: succinyl-CoA; SUC: succinate; FMA: fumarate; MAL: malate; OAA: oxaloacetate
| [1] |
Vallée M, Vitiello S, Bellocchio L, et al. Pregnenolone can protect the brain from cannabis intoxication[J]. Science, 2014, 343(6166):94-98.
doi: 10.1126/science.1243985 URL |
| [2] | 熊亮斌. 分枝杆菌甾醇代谢途径的解析及高产甾体医药中间体工程菌株的构建[D]. 上海:华东理工大学, 2017. |
| Xiong LB. Analysis of the sterol metabolic pathway in mycobacteria and the modification of high-yield steroidal pharmaceutical precursors producing strains[D]. Shanghai:East China University of Science and Technology, 2017. | |
| [3] | Donova MV, Dovbnya DV, Sukhodolskaya GV, et al. Microbial conversion of sterol-containing soybean oil production waste[J]. Journal of Chemical Technology and Biotechnology, 2005, 80(1):55-60. |
| [4] |
Yao K, Xu LQ, Wang FQ, et al. Characterization and engineering of 3-ketosteroid-Δ1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3, 17-dione through the catabolism of sterols[J]. Metabolic Engineering, 2014, 24:181-191.
doi: 10.1016/j.ymben.2014.05.005 URL |
| [5] | Liu HH, Xu LQ, Yao K, et al. Characterization and engineering of 3-ketosteroid 9α-hydroxylases in Mycobacterium neoaurum ATCC 25795 for the development of androst-1, 4-diene-3, 17-dione and 9α-hydroxy-androst-4-ene-3, 17-dione producing strains[J]. Applied and Environmental Microbiology, 2018, 84(14):e-02777-02717. |
| [6] |
Xu LQ, Liu YJ, Yao K, et al. Unraveling and engineering the production of 23, 24-bisnorcholenic steroids in sterol metabolism[J]. Scientific Reports, 2016, 6:21928.
doi: 10.1038/srep21928 pmid: 26898409 |
| [7] |
Donova MV, Egorova OV. Microbial steroid transformations:current state and prospects[J]. Applied Microbiology and Biotechnology, 2012, 94(6):1423-1447.
doi: 10.1007/s00253-012-4078-0 URL |
| [8] |
Fernández-Cabezón L, Galán B, García JL. Unravelling a new catabolic pathway of C-19 steroids in Mycobacterium smegmatis[J]. Environmental Microbiology, 2018, 20(5):1815-1827.
doi: 10.1111/1462-2920.14114 pmid: 29611894 |
| [9] |
Wipperman MF, Sampson NS, Thomas ST. Pathogen roid rage:cholesterol utilization by Mycobacterium tuberculosis[J]. Critical Reviews in Biochemistry and Molecular Biology, 2014, 49(4):269-293.
doi: 10.3109/10409238.2014.895700 pmid: 24611808 |
| [10] |
Liu M, Xiong LB, Tao X, et al. Metabolic adaptation of Mycobacterium neoaurum ATCC 25795 in the catabolism of sterols for producing important steroid intermediates[J]. Journal of Agricultural and Food Chemistry, 2018, 66(45):12141-12150.
doi: 10.1021/acs.jafc.8b04777 URL |
| [11] |
Xiong LB, Liu HH, Xu LQ, et al. Role identification and application of SigD in the transformation of soybean phytosterol to 9α-hydroxy-4-androstene-3, 17-dione in Mycobacterium neoaurum[J]. Journal of Agricultural and Food Chemistry, 2017, 65(3):626-631.
doi: 10.1021/acs.jafc.6b05314 URL |
| [12] |
Xiong LB, Sun WJ, Liu YJ, et al. Enhancement of 9α-hydroxy-4-androstene-3, 17-dione production from soybean phytosterols by deficiency of a regulated intramembrane proteolysis metalloprotease in Mycobacterium neoaurum[J]. Journal of Agricultural and Food Chemistry, 2017, 65(48):10520-10525.
doi: 10.1021/acs.jafc.7b03766 URL |
| [13] |
Yao K, Wang FQ, Zhang HC, et al. Identification and engineering of cholesterol oxidases involved in the initial step of sterols catabolism in Mycobacterium neoaurum[J]. Metabolic Engineering, 2013, 15:75-87.
doi: 10.1016/j.ymben.2012.10.005 URL |
| [14] |
Gonzalo-Asensio J, Malaga W, Pawlik A, et al. Evolutionary history of tuberculosis shaped by conserved mutations in the PhoPR virulence regulator[J]. Proceedings of the National Academy of Sciences USA, 2014, 111(31):11491-11496.
doi: 10.1073/pnas.1406693111 URL |
| [15] | Broset E, Martín C, Gonzalo-Asensio J. Evolutionary landscape of the Mycobacterium tuberculosis complex from the viewpoint of PhoPR:implications for virulence regulation and application to vaccine development[J]. Mbio, 2015, 6(5):e01289. |
| [16] |
Xiong LB, Liu HH, Zhao M, et al. Enhancing the bioconversion of phytosterols to steroidal intermediates by the deficiency of kasB in the cell wall synjournal of Mycobacterium neoaurum[J]. Microbial Cell Factories, 2020, 19:80.
doi: 10.1186/s12934-020-01335-y URL |
| [17] |
Xiong LB, Liu HH, Song XW, et al. Improving the biotransformation of phytosterols to 9α-hydroxy-4-androstene-3, 17-dione by deleting embC associated with the assembly of cell envelope in Mycobacterium neoaurum[J]. Journal of Biotechnology, 2020, 323(2020):341-346.
doi: 10.1016/j.jbiotec.2020.09.019 URL |
| [18] |
Baker JJ, Johnson BK, Abramovitch RB. Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources[J]. Molecular Microbiology, 2014, 94(1):56-69.
doi: 10.1111/mmi.2014.94.issue-1 URL |
| [19] |
Xiong LB, Liu HH, Xu LQ, et al. Improving the production of 22-hydroxy-23, 24-bisnorchol-4-ene-3-one from sterols in Mycobacterium neoaurum by increasing cell permeability and modifying multiple genes[J]. Microbial Cell Factories, 2017, 16:89.
doi: 10.1186/s12934-017-0705-x URL |
| [1] | DONG Cong, GAO Qing-hua, WANG Yue, LUO Tong-yang, WANG Qing-qing. Increasing the Expression of FAD-dependent Glucose Dehydrogenase by Recombinant Pichia pastoris Using a Combined Strategy [J]. Biotechnology Bulletin, 2023, 39(6): 316-324. |
| [2] | NIU Yu-hui, LI Xiang-rong, WU Bei, LI Hong-shan, LI Dian-yu, CHEN Lei, WEI Suo-cheng, FENG Ruo-fei. Effects of Glucose and Sodium Butyrate on the rHSA Yield in CHO-rHSA Engineering Cell Line [J]. Biotechnology Bulletin, 2022, 38(7): 278-286. |
| [3] | LIAO Zhao-min, CAI Jun, LIN Jian-guo, DU Xin, WANG Chang-gao. Expression of Glucose Oxidase Gene from Aspergillus niger in Pichia pastoris and Optimization of Enzyme Production Conditions [J]. Biotechnology Bulletin, 2021, 37(6): 97-107. |
| [4] | HUO Ming-yue, ZHANG Ge, SU Yu-long, LI Xing, ZHANG Hai-bo, ZHENG Rong, LIU Hao-bao. Isolation of an Acetoin-producing Strain Resistant to High Concentration Glucose and Identification of Metabolites [J]. Biotechnology Bulletin, 2020, 36(8): 53-60. |
| [5] | CHEN Qiao, WU Hai-ying, WANG Zong-shou, XIE Yu-kang, LI Yi-qing, SUN Jun-song. Multiple-site Mutations in Escherichia coli Capable of High-density Growing Induced from the Biosynthesis of Polyhydroxybutyrate [J]. Biotechnology Bulletin, 2020, 36(7): 112-118. |
| [6] | DONG Cong, GAO Qing-hua, WANG Yue, LUO Tong-yang. Expression and Enzymatic Characterization of Codon-optimized FAD-dependent Glucose Dehydrogenase in Pichia pastoris [J]. Biotechnology Bulletin, 2019, 35(7): 114-120. |
| [7] | LI Ji-xuan, YU Lei, LI Jing-mei, ZHENG Gui-lan, WANG Hong-zhong. Construction of Co-expression System of S-imine Reductase and Glucose Dehydrogenase and Synthesis of Chiral Amine [J]. Biotechnology Bulletin, 2019, 35(1): 105-111. |
| [8] | GAO Qing-hua, DONG Cong, WANG Yue, HU Mei-rong, WANG Qing-qing, WANG Yun-peng, LUO Tong-yang, LIU Lei. Enhancement of Glucose Oxidase in Pichia pastoris by Co-expressing Chaperone PDI and Ero1 [J]. Biotechnology Bulletin, 2018, 34(7): 174-179. |
| [9] | DAI Chen-xia, CAO Hui-fang, LUO Hui-ying, YAO Bin, BAI Ying-guo, XU Bo. Cloning,Expression and Conversion Application of Glucose Isomerase from Alicyclobacillus sp.A4 [J]. Biotechnology Bulletin, 2018, 34(11): 144-151. |
| [10] | MA Shi-nan, LI Zhong-yao, GUO Xing-rong. Study on the Molecular Mechanism of ANGPTL8 in Regulating the Glucose Metabolism [J]. Biotechnology Bulletin, 2017, 33(11): 200-206. |
| [11] | GAO Qing-hua, HU Mei-rong, WU Fang-tong, TAO Yong, WANG Yun-peng, LUO Tong-yang, HU Chang-ying. Cloning of Gene for a Glucose Oxidase from Penicillium notatum and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2016, 32(7): 152-159. |
| [12] | Lin Shifeng, Fu Qiang, Yu Jing, Zhao Jiehong, Ren Xueliang, Wang Rengang. Cloning and Expression Analysis of Cytosolic 6-phosphogluconate Dehydrogenase Gene in Tobacco(Nicotiana tabacum) [J]. Biotechnology Bulletin, 2015, 31(5): 113-119. |
| [13] | Guo Wenyi, Sun Qinghui, Song Lina, Gao Songsong, Yang Hongjiang. Breeding and Fermentation Characterization of Mutants of Bacillus licheniformis for 2,3-Butanediol Production [J]. Biotechnology Bulletin, 2014, 0(8): 159-163. |
| [14] | Wang Shuaikun Hao Jieqing Wang Zhenwei Shi Hui Meng Yanfa. Studies on Fermentation Conditions for r-GOD Production by Pichia pastoris GS115 [J]. Biotechnology Bulletin, 2013, 0(9): 136-141. |
| [15] | Liu Yu, Li Piwu. Review on High-level Aspergillus niger Glucose Oxidase Production Engineering Strains [J]. Biotechnology Bulletin, 2013, 0(7): 12-19. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||