








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (8): 65-74.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0173
Previous Articles Next Articles
LI Qi1( ), WANG Yi-chao2, LIU Chang1, TAN He-xin1(
), WANG Yi-chao2, LIU Chang1, TAN He-xin1( )
)
Received:2021-02-09
Online:2021-08-26
Published:2021-09-10
Contact:
TAN He-xin
E-mail:liqi0121@163.com;hexintan@163.com
LI Qi, WANG Yi-chao, LIU Chang, TAN He-xin. Genome-wide Identification and Bioinformatics Analysis of R2R3-MYB Transcription Factors in Artemisia annua[J]. Biotechnology Bulletin, 2021, 37(8): 65-74.
| Protein code | MYB name | Protein code | MYB name | Protein code | MYB name |
|---|---|---|---|---|---|
| PWA71644.1 | AaMYB2R1 | PWA99792.1 | AaMYB2R45 | PWA38419.1 | AaMYB2R89 |
| PWA43204.1 | AaMYB2R2 | PWA41205.1 | AaMYB2R46 | PWA56935.1 | AaMYB2R90 |
| PWA86000.1 | AaMYB2R3 | PWA96412.1 | AaMYB2R47 | PWA35205.1 | AaMYB2R91 |
| PWA65390.1 | AaMYB2R4 | PWA50143.1 | AaMYB2R48 | PWA76413.1 | AaMYB2R92 |
| PWA47067.1 | AaMYB2R5 | PWA95387.1 | AaMYB2R49 | PWA61358.1 | AaMYB2R93 |
| PWA39356.1 | AaMYB2R6 | PWA87312.1 | AaMYB2R50 | PWA68967.1 | AaMYB2R94 |
| PWA75916.1 | AaMYB2R7 | PWA72501.1 | AaMYB2R51 | PWA70735.1 | AaMYB2R95 |
| PWA43649.1 | AaMYB2R8 | PWA88612.1 | AaMYB2R52 | PWA65469.1 | AaMYB2R96 |
| PWA51649.1 | AaMYB2R9 | PWA71899.1 | AaMYB2R53 | PWA73667.1 | AaMYB2R97 |
| PWA64888.1 | AaMYB2R10 | PWA60948.1 | AaMYB2R54 | PWA60339.1 | AaMYB2R98 |
| PWA67214.1 | AaMYB2R11 | PWA66225.1 | AaMYB2R55 | PWA47768.1 | AaMYB2R99 |
| PWA89491.1 | AaMYB2R12 | PWA35659.1 | AaMYB2R56 | PWA65296.1 | AaMYB2R100 |
| PWA46461.1 | AaMYB2R13 | PWA98977.1 | AaMYB2R57 | PWA93717.1 | AaMYB2R101 |
| PWA78544.1 | AaMYB2R14 | PWA87993.1 | AaMYB2R58 | PWA99549.1 | AaMYB2R102 |
| PWA71245.1 | AaMYB2R15 | PWA63153.1 | AaMYB2R59 | PWA44719.1 | AaMYB2R103 |
| PWA96367.1 | AaMYB2R16 | PWA70731.1 | AaMYB2R60 | PWA74420.1 | AaMYB2R104 |
| PWA52706.1 | AaMYB2R17 | PWA90922.1 | AaMYB2R61 | PWA74110.1 | AaMYB2R105 |
| PWA64358.1 | AaMYB2R18 | PWA56311.1 | AaMYB2R62 | PWA66731.1 | AaMYB2R106 |
| PWA52104.1 | AaMYB2R19 | PWA81663.1 | AaMYB2R63 | PWA46401.1 | AaMYB2R107 |
| PWA34289.1 | AaMYB2R20 | PWA75167.1 | AaMYB2R64 | PWA60446.1 | AaMYB2R108 |
| PWA45135.1 | AaMYB2R21 | PWA64917.1 | AaMYB2R65 | PWA50364.1 | AaMYB2R109 |
| PWA46713.1 | AaMYB2R22 | PWA54615.1 | AaMYB2R66 | PWA81967.1 | AaMYB2R110 |
| PWA60682.1 | AaMYB2R23 | PWA50450.1 | AaMYB2R67 | PWA55616.1 | AaMYB2R111 |
| PWA52704.1 | AaMYB2R24 | PWA55247.1 | AaMYB2R68 | PWA39801.1 | AaMYB2R112 |
| PWA86815.1 | AaMYB2R25 | PWA78936.1 | AaMYB2R69 | PWA65860.1 | AaMYB2R113 |
| PWA52975.1 | AaMYB2R26 | PWA48003.1 | AaMYB2R70 | PWA93789.1 | AaMYB2R114 |
| PWA84715.1 | AaMYB2R27 | PWA84612.1 | AaMYB2R71 | PWA70975.1 | AaMYB2R115 |
| PWA47058.1 | AaMYB2R28 | PWA98963.1 | AaMYB2R72 | PWA63331.1 | AaMYB2R116 |
| PWA48985.1 | AaMYB2R29 | PWA85713.1 | AaMYB2R73 | PWA37853.1 | AaMYB2R117 |
| PWA71307.1 | AaMYB2R30 | PWA67086.1 | AaMYB2R74 | PWA78715.1 | AaMYB2R118 |
| PWA66224.1 | AaMYB2R31 | PWA47522.1 | AaMYB2R75 | PWA87301.1 | AaMYB2R119 |
| PWA66489.1 | AaMYB2R32 | PWA96303.1 | AaMYB2R76 | PWA62998.1 | AaMYB2R120 |
| PWA88884.1 | AaMYB2R33 | PWA74168.1 | AaMYB2R77 | PWA94056.1 | AaMYB2R121 |
| PWA60694.1 | AaMYB2R34 | PWA80451.1 | AaMYB2R78 | PWA35582.1 | AaMYB2R122 |
| PWA96157.1 | AaMYB2R35 | PWA60300.1 | AaMYB2R79 | PWA92852.1 | AaMYB2R123 |
| PWA60193.1 | AaMYB2R36 | PWA72588.1 | AaMYB2R80 | PWA56697.1 | AaMYB2R124 |
| PWA89756.1 | AaMYB2R37 | PWA60276.1 | AaMYB2R81 | PWA93922.1 | AaMYB2R125 |
| PWA89743.1 | AaMYB2R38 | PWA77673.1 | AaMYB2R82 | PWA82717.1 | AaMYB2R126 |
| PWA59700.1 | AaMYB2R39 | PWA70733.1 | AaMYB2R83 | PWA38555.1 | AaMYB2R127 |
| PWA72169.1 | AaMYB2R40 | PWA56370.1 | AaMYB2R84 | PWA43034.1 | AaMYB2R128 |
| PWA34017.1 | AaMYB2R41 | PWA50987.1 | AaMYB2R85 | PWA70165.1 | AaMYB2R129 |
| PWA87210.1 | AaMYB2R42 | PWA88000.1 | AaMYB2R86 | PWA95682.1 | AaMYB2R130 |
| PWA71841.1 | AaMYB2R43 | PWA85391.1 | AaMYB2R87 | PWA89358.1 | AaMYB2R131 |
| PWA79178.1 | AaMYB2R44 | PWA71534.1 | AaMYB2R88 | PWA90842.1 | AaMYB2R132 |
Table 1 Screening result of R2R3-MYB transcription factors in A. annua
| Protein code | MYB name | Protein code | MYB name | Protein code | MYB name |
|---|---|---|---|---|---|
| PWA71644.1 | AaMYB2R1 | PWA99792.1 | AaMYB2R45 | PWA38419.1 | AaMYB2R89 |
| PWA43204.1 | AaMYB2R2 | PWA41205.1 | AaMYB2R46 | PWA56935.1 | AaMYB2R90 |
| PWA86000.1 | AaMYB2R3 | PWA96412.1 | AaMYB2R47 | PWA35205.1 | AaMYB2R91 |
| PWA65390.1 | AaMYB2R4 | PWA50143.1 | AaMYB2R48 | PWA76413.1 | AaMYB2R92 |
| PWA47067.1 | AaMYB2R5 | PWA95387.1 | AaMYB2R49 | PWA61358.1 | AaMYB2R93 |
| PWA39356.1 | AaMYB2R6 | PWA87312.1 | AaMYB2R50 | PWA68967.1 | AaMYB2R94 |
| PWA75916.1 | AaMYB2R7 | PWA72501.1 | AaMYB2R51 | PWA70735.1 | AaMYB2R95 |
| PWA43649.1 | AaMYB2R8 | PWA88612.1 | AaMYB2R52 | PWA65469.1 | AaMYB2R96 |
| PWA51649.1 | AaMYB2R9 | PWA71899.1 | AaMYB2R53 | PWA73667.1 | AaMYB2R97 |
| PWA64888.1 | AaMYB2R10 | PWA60948.1 | AaMYB2R54 | PWA60339.1 | AaMYB2R98 |
| PWA67214.1 | AaMYB2R11 | PWA66225.1 | AaMYB2R55 | PWA47768.1 | AaMYB2R99 |
| PWA89491.1 | AaMYB2R12 | PWA35659.1 | AaMYB2R56 | PWA65296.1 | AaMYB2R100 |
| PWA46461.1 | AaMYB2R13 | PWA98977.1 | AaMYB2R57 | PWA93717.1 | AaMYB2R101 |
| PWA78544.1 | AaMYB2R14 | PWA87993.1 | AaMYB2R58 | PWA99549.1 | AaMYB2R102 |
| PWA71245.1 | AaMYB2R15 | PWA63153.1 | AaMYB2R59 | PWA44719.1 | AaMYB2R103 |
| PWA96367.1 | AaMYB2R16 | PWA70731.1 | AaMYB2R60 | PWA74420.1 | AaMYB2R104 |
| PWA52706.1 | AaMYB2R17 | PWA90922.1 | AaMYB2R61 | PWA74110.1 | AaMYB2R105 |
| PWA64358.1 | AaMYB2R18 | PWA56311.1 | AaMYB2R62 | PWA66731.1 | AaMYB2R106 |
| PWA52104.1 | AaMYB2R19 | PWA81663.1 | AaMYB2R63 | PWA46401.1 | AaMYB2R107 |
| PWA34289.1 | AaMYB2R20 | PWA75167.1 | AaMYB2R64 | PWA60446.1 | AaMYB2R108 |
| PWA45135.1 | AaMYB2R21 | PWA64917.1 | AaMYB2R65 | PWA50364.1 | AaMYB2R109 |
| PWA46713.1 | AaMYB2R22 | PWA54615.1 | AaMYB2R66 | PWA81967.1 | AaMYB2R110 |
| PWA60682.1 | AaMYB2R23 | PWA50450.1 | AaMYB2R67 | PWA55616.1 | AaMYB2R111 |
| PWA52704.1 | AaMYB2R24 | PWA55247.1 | AaMYB2R68 | PWA39801.1 | AaMYB2R112 |
| PWA86815.1 | AaMYB2R25 | PWA78936.1 | AaMYB2R69 | PWA65860.1 | AaMYB2R113 |
| PWA52975.1 | AaMYB2R26 | PWA48003.1 | AaMYB2R70 | PWA93789.1 | AaMYB2R114 |
| PWA84715.1 | AaMYB2R27 | PWA84612.1 | AaMYB2R71 | PWA70975.1 | AaMYB2R115 |
| PWA47058.1 | AaMYB2R28 | PWA98963.1 | AaMYB2R72 | PWA63331.1 | AaMYB2R116 |
| PWA48985.1 | AaMYB2R29 | PWA85713.1 | AaMYB2R73 | PWA37853.1 | AaMYB2R117 |
| PWA71307.1 | AaMYB2R30 | PWA67086.1 | AaMYB2R74 | PWA78715.1 | AaMYB2R118 |
| PWA66224.1 | AaMYB2R31 | PWA47522.1 | AaMYB2R75 | PWA87301.1 | AaMYB2R119 |
| PWA66489.1 | AaMYB2R32 | PWA96303.1 | AaMYB2R76 | PWA62998.1 | AaMYB2R120 |
| PWA88884.1 | AaMYB2R33 | PWA74168.1 | AaMYB2R77 | PWA94056.1 | AaMYB2R121 |
| PWA60694.1 | AaMYB2R34 | PWA80451.1 | AaMYB2R78 | PWA35582.1 | AaMYB2R122 |
| PWA96157.1 | AaMYB2R35 | PWA60300.1 | AaMYB2R79 | PWA92852.1 | AaMYB2R123 |
| PWA60193.1 | AaMYB2R36 | PWA72588.1 | AaMYB2R80 | PWA56697.1 | AaMYB2R124 |
| PWA89756.1 | AaMYB2R37 | PWA60276.1 | AaMYB2R81 | PWA93922.1 | AaMYB2R125 |
| PWA89743.1 | AaMYB2R38 | PWA77673.1 | AaMYB2R82 | PWA82717.1 | AaMYB2R126 |
| PWA59700.1 | AaMYB2R39 | PWA70733.1 | AaMYB2R83 | PWA38555.1 | AaMYB2R127 |
| PWA72169.1 | AaMYB2R40 | PWA56370.1 | AaMYB2R84 | PWA43034.1 | AaMYB2R128 |
| PWA34017.1 | AaMYB2R41 | PWA50987.1 | AaMYB2R85 | PWA70165.1 | AaMYB2R129 |
| PWA87210.1 | AaMYB2R42 | PWA88000.1 | AaMYB2R86 | PWA95682.1 | AaMYB2R130 |
| PWA71841.1 | AaMYB2R43 | PWA85391.1 | AaMYB2R87 | PWA89358.1 | AaMYB2R131 |
| PWA79178.1 | AaMYB2R44 | PWA71534.1 | AaMYB2R88 | PWA90842.1 | AaMYB2R132 |
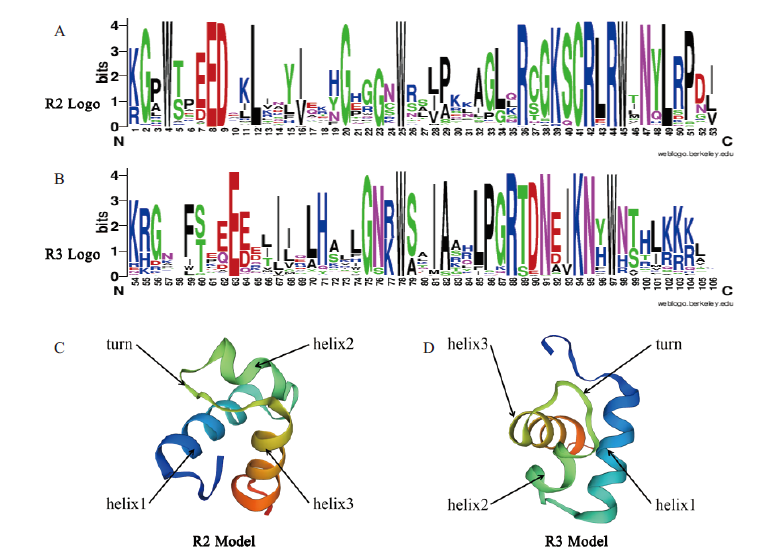
Fig.1 Conserved domain analysis result of R2R3-MYB transcription factors in A. annua A:Analysis result of R2 domain by Weblogo. B:Analysis result of R3 domain. C:Predicted structure of R2 domain by Swiss-Model.D:Predicted structure of R3 domain by Swiss-Model
| Motif | Top scoring sequence | E-value | Site |
|---|---|---|---|
| 1 | AGLQRCGKSCRLRWTNYLRPDJKRGNFTEEEEELIJQLHALLGNRWSAIA | 1.3e-4561 | 125 |
| 2 | HLPGRTDNEIKNYWNTHLKKK | 1.3e-1932 | 126 |
| 3 | KKGPWTPEEDQKLINYIZKHGHGN | 3.0e-1621 | 127 |
| 4 | MGRAPCCDKNG | 8.1e-356 | 60 |
| 5 | LLKMGIDPVTHKPK | 2.80E-228 | 37 |
| 6 | TNLSHMAQWENARLEAEARLVRQSRVVS | 6.00E-132 | 12 |
| 7 | WRSLPK | 3.00E-129 | 110 |
| 8 | INKSSVSSLLVPQPLCFDVLKAWQGJTLSKLSSL | 7.90E-35 | 5 |
| 9 | TMSKGQWERRLQTDIHMAKQALCDALSIDNKSI | 2.00E-32 | 4 |
| 10 | LDLSDLSSJLNSAIENLSNLLNLQSLVNP | 1.10E-30 | 7 |
| 11 | EFVDVMQEMIAKEVRNYI | 2.70E-29 | 7 |
| 12 | QLKCDVNSKQFKDTMRYLWMPRLLEKIZA | 4.40E-28 | 5 |
| 13 | SDLTDCYYPISESQNQNFLQINNTVGDEFDCPIISPTIYFNREMDFEGQM | 2.60E-27 | 3 |
| 14 | PLGLVNDLISQASCQDMIMDDDLLWSTJD | 2.80E-25 | 8 |
| 15 | NVVESVEPQTLKGMSTEYHMN | 9.10E-19 | 4 |
| 16 | KEEATQHTLEVNDMIYQDNYLAEAFTNHLVNDANCHGHLED | 6.80E-22 | 4 |
| 17 | YASSAENIARLLPNWMKSSSKSSQTSS | 8.70E-17 | 5 |
| 18 | PPSSYEPAGNFWTEPFLVDN | 5.90E-16 | 10 |
| 19 | FNSWDECATWLLDCQDFGIHDFGLDCFGZVEM | 1.50E-09 | 2 |
| 20 | KQKLGTKDSEPLQQHAKVSVIKPQPQTFSRIQNC | 6.40E-10 | 3 |
| 21 | DSIFSTPISSPTPLNSSSTLINSSSTEDERESYC | 2.20E-09 | 4 |
| 22 | TMWPCKLNPIQEKJMQNLQLLNESSNPLMMQHFSPSTPQKIEYY | 2.40E-19 | 3 |
| 23 | CSSMTNEEFNDFAVQQPLLKRSVSDGSGVP | 9.70E-13 | 3 |
| 24 | PGTIDDLQYFWDQLCPVENLELRNNHLD | 1.00E-07 | 3 |
| 25 | MAQDVARWWDTVSEDGLWGGFLWNQENDHPNKQAIEQSFSPCF | 1.30E-07 | 2 |
| 26 | EIEGFKDMDGAKDDMTWWSBEFDANSASSNSWDSBGVLDH | 1.10E-06 | 3 |
| 27 | TDDGVNNALDIPQGLISSPIESGDKIDEYLDELFNDEENEFEGEIDWTLD | 2.00E-06 | 2 |
| 28 | NHFNEDIDGKIKVEEVVGFENHWPGENFKIGEWDFEGFLANIPSJPYLDF | 2.10E-06 | 2 |
| 29 | FKFEIPESLEFDDFM | 5.70E-06 | 6 |
| 30 | DYFLRVWHSEVGESFR | 7.50E-05 | 4 |
| 31 | SSSETSSSSLSGSDYVVPSDV | 1.70E-04 | 7 |
| 32 | KRRVGRVSRRVAKKYNKDRV | 2.40E-04 | 3 |
| 33 | GMFMSMHDHDJMAICME | 2.90E-04 | 2 |
| 34 | FTKRKCRSKWPKPSPILQNYIKSLNLPRG | 4.80E-04 | 2 |
| 35 | ELQQFLAFQAEIDEFLS | 3.00E-05 | 3 |
| 36 | PFIDFLGVG | 4.90E-05 | 5 |
| 37 | VSVNDPPTSLSLSLPGV | 4.90E-04 | 7 |
| 38 | DSSDTQLQLFFDFPGPNDANF | 6.30E-04 | 4 |
| 39 | SGGEEDDQMNMMMD | 1.60E-03 | 4 |
| 40 | AAQVNKLPKILFADWLSLDEFH | 1.70E-03 | 3 |
| 41 | DSNYWSNILDKLGN | 3.50E-03 | 4 |
| 42 | YTRDNSSNTAVSPV | 1.30E-02 | 3 |
| 43 | WNMDDLWQFR | 4.20E-02 | 4 |
| 44 | FHLPDEVMWDF | 9.90E-02 | 3 |
| 45 | YDHGHHHHHHN | 4.50E-02 | 3 |
Table 2 Domain search result of R2R3-MYB transcription factors in Artemisia annua by MEME
| Motif | Top scoring sequence | E-value | Site |
|---|---|---|---|
| 1 | AGLQRCGKSCRLRWTNYLRPDJKRGNFTEEEEELIJQLHALLGNRWSAIA | 1.3e-4561 | 125 |
| 2 | HLPGRTDNEIKNYWNTHLKKK | 1.3e-1932 | 126 |
| 3 | KKGPWTPEEDQKLINYIZKHGHGN | 3.0e-1621 | 127 |
| 4 | MGRAPCCDKNG | 8.1e-356 | 60 |
| 5 | LLKMGIDPVTHKPK | 2.80E-228 | 37 |
| 6 | TNLSHMAQWENARLEAEARLVRQSRVVS | 6.00E-132 | 12 |
| 7 | WRSLPK | 3.00E-129 | 110 |
| 8 | INKSSVSSLLVPQPLCFDVLKAWQGJTLSKLSSL | 7.90E-35 | 5 |
| 9 | TMSKGQWERRLQTDIHMAKQALCDALSIDNKSI | 2.00E-32 | 4 |
| 10 | LDLSDLSSJLNSAIENLSNLLNLQSLVNP | 1.10E-30 | 7 |
| 11 | EFVDVMQEMIAKEVRNYI | 2.70E-29 | 7 |
| 12 | QLKCDVNSKQFKDTMRYLWMPRLLEKIZA | 4.40E-28 | 5 |
| 13 | SDLTDCYYPISESQNQNFLQINNTVGDEFDCPIISPTIYFNREMDFEGQM | 2.60E-27 | 3 |
| 14 | PLGLVNDLISQASCQDMIMDDDLLWSTJD | 2.80E-25 | 8 |
| 15 | NVVESVEPQTLKGMSTEYHMN | 9.10E-19 | 4 |
| 16 | KEEATQHTLEVNDMIYQDNYLAEAFTNHLVNDANCHGHLED | 6.80E-22 | 4 |
| 17 | YASSAENIARLLPNWMKSSSKSSQTSS | 8.70E-17 | 5 |
| 18 | PPSSYEPAGNFWTEPFLVDN | 5.90E-16 | 10 |
| 19 | FNSWDECATWLLDCQDFGIHDFGLDCFGZVEM | 1.50E-09 | 2 |
| 20 | KQKLGTKDSEPLQQHAKVSVIKPQPQTFSRIQNC | 6.40E-10 | 3 |
| 21 | DSIFSTPISSPTPLNSSSTLINSSSTEDERESYC | 2.20E-09 | 4 |
| 22 | TMWPCKLNPIQEKJMQNLQLLNESSNPLMMQHFSPSTPQKIEYY | 2.40E-19 | 3 |
| 23 | CSSMTNEEFNDFAVQQPLLKRSVSDGSGVP | 9.70E-13 | 3 |
| 24 | PGTIDDLQYFWDQLCPVENLELRNNHLD | 1.00E-07 | 3 |
| 25 | MAQDVARWWDTVSEDGLWGGFLWNQENDHPNKQAIEQSFSPCF | 1.30E-07 | 2 |
| 26 | EIEGFKDMDGAKDDMTWWSBEFDANSASSNSWDSBGVLDH | 1.10E-06 | 3 |
| 27 | TDDGVNNALDIPQGLISSPIESGDKIDEYLDELFNDEENEFEGEIDWTLD | 2.00E-06 | 2 |
| 28 | NHFNEDIDGKIKVEEVVGFENHWPGENFKIGEWDFEGFLANIPSJPYLDF | 2.10E-06 | 2 |
| 29 | FKFEIPESLEFDDFM | 5.70E-06 | 6 |
| 30 | DYFLRVWHSEVGESFR | 7.50E-05 | 4 |
| 31 | SSSETSSSSLSGSDYVVPSDV | 1.70E-04 | 7 |
| 32 | KRRVGRVSRRVAKKYNKDRV | 2.40E-04 | 3 |
| 33 | GMFMSMHDHDJMAICME | 2.90E-04 | 2 |
| 34 | FTKRKCRSKWPKPSPILQNYIKSLNLPRG | 4.80E-04 | 2 |
| 35 | ELQQFLAFQAEIDEFLS | 3.00E-05 | 3 |
| 36 | PFIDFLGVG | 4.90E-05 | 5 |
| 37 | VSVNDPPTSLSLSLPGV | 4.90E-04 | 7 |
| 38 | DSSDTQLQLFFDFPGPNDANF | 6.30E-04 | 4 |
| 39 | SGGEEDDQMNMMMD | 1.60E-03 | 4 |
| 40 | AAQVNKLPKILFADWLSLDEFH | 1.70E-03 | 3 |
| 41 | DSNYWSNILDKLGN | 3.50E-03 | 4 |
| 42 | YTRDNSSNTAVSPV | 1.30E-02 | 3 |
| 43 | WNMDDLWQFR | 4.20E-02 | 4 |
| 44 | FHLPDEVMWDF | 9.90E-02 | 3 |
| 45 | YDHGHHHHHHN | 4.50E-02 | 3 |

Fig.2 Phylogenetic tree of R2R3-MYB transcription factors in A. annua and Arabidopsis thaliana ○ Sequence of A. annua,● Sequence of Arabidopsis thaliana
| GO term | Ontology | Description | Gene number |
|---|---|---|---|
| GO:0005634 | C | Nucleus | 128 |
| GO:0043565 | F | Sequence-specific DNA binding | 95 |
| GO:0044212 | F | Transcription regulatory region DNA binding | 84 |
| GO:0003700 | F | DNA-binding transcription factor activity | 83 |
| GO:0030154 | P | Cell differentiation | 80 |
| GO:0006355 | P | Regulation of transcription,DNA-templated | 67 |
| GO:0003677 | F | DNA binding | 36 |
| GO:0009733 | P | Response to auxin | 4 |
| GO:0010119 | P | Regulation of stomatal movement | 3 |
| GO:0009751 | P | Response to salicylic acid | 2 |
| GO:0048658 | P | Anther wall tapetum development | 2 |
| GO:0005515 | F | Protein binding | 1 |
| GO:0005524 | F | ATP binding | 1 |
| GO:0016021 | C | Integral component of membrane | 1 |
| GO:0009739 | P | Response to gibberellin | 1 |
| GO:0009753 | P | Response to jasmonic acid | 1 |
| GO:0048481 | P | Plant ovule development | 1 |
| GO:0090377 | P | Seed trichome initiation | 1 |
| GO:0004222 | F | Metalloendopeptidase activity | 1 |
| GO:0006508 | P | Proteolysis | 1 |
| GO:0003824 | F | Catalytic activity | 1 |
| GO:0001101 | P | Response to acid chemical | 1 |
| GO:1901700 | P | Response to oxygen-containing compound | 1 |
| GO:0052545 | P | Callose localization | 1 |
| GO:0055046 | P | Microgametogenesis | 1 |
| GO:0050789 | P | Regulation of biological process | 1 |
| GO:0010090 | P | Trichome morphogenesis | 1 |
| GO:0048354 | P | Mucilage biosynthetic process involved in seed coat development | 1 |
| GO:0048522 | P | Positive regulation of cellular process | 1 |
| GO:0048523 | P | Negative regulation of cellular process | 1 |
| GO:0050793 | P | Regulation of developmental process | 1 |
| GO:0044839 | P | Cell cycle G2/M phase transition | 1 |
| GO:0045893 | P | Positive regulation of transcription,DNA-templated | 1 |
| GO:0048235 | P | Pollen sperm cell differentiation | 1 |
| GO:0048555 | C | Generative cell nucleus | 1 |
| GO:0055047 | P | Generative cell mitosis | 1 |
Table 3 GO annotation count result of R2R3-MYB transcription factors in A. annua
| GO term | Ontology | Description | Gene number |
|---|---|---|---|
| GO:0005634 | C | Nucleus | 128 |
| GO:0043565 | F | Sequence-specific DNA binding | 95 |
| GO:0044212 | F | Transcription regulatory region DNA binding | 84 |
| GO:0003700 | F | DNA-binding transcription factor activity | 83 |
| GO:0030154 | P | Cell differentiation | 80 |
| GO:0006355 | P | Regulation of transcription,DNA-templated | 67 |
| GO:0003677 | F | DNA binding | 36 |
| GO:0009733 | P | Response to auxin | 4 |
| GO:0010119 | P | Regulation of stomatal movement | 3 |
| GO:0009751 | P | Response to salicylic acid | 2 |
| GO:0048658 | P | Anther wall tapetum development | 2 |
| GO:0005515 | F | Protein binding | 1 |
| GO:0005524 | F | ATP binding | 1 |
| GO:0016021 | C | Integral component of membrane | 1 |
| GO:0009739 | P | Response to gibberellin | 1 |
| GO:0009753 | P | Response to jasmonic acid | 1 |
| GO:0048481 | P | Plant ovule development | 1 |
| GO:0090377 | P | Seed trichome initiation | 1 |
| GO:0004222 | F | Metalloendopeptidase activity | 1 |
| GO:0006508 | P | Proteolysis | 1 |
| GO:0003824 | F | Catalytic activity | 1 |
| GO:0001101 | P | Response to acid chemical | 1 |
| GO:1901700 | P | Response to oxygen-containing compound | 1 |
| GO:0052545 | P | Callose localization | 1 |
| GO:0055046 | P | Microgametogenesis | 1 |
| GO:0050789 | P | Regulation of biological process | 1 |
| GO:0010090 | P | Trichome morphogenesis | 1 |
| GO:0048354 | P | Mucilage biosynthetic process involved in seed coat development | 1 |
| GO:0048522 | P | Positive regulation of cellular process | 1 |
| GO:0048523 | P | Negative regulation of cellular process | 1 |
| GO:0050793 | P | Regulation of developmental process | 1 |
| GO:0044839 | P | Cell cycle G2/M phase transition | 1 |
| GO:0045893 | P | Positive regulation of transcription,DNA-templated | 1 |
| GO:0048235 | P | Pollen sperm cell differentiation | 1 |
| GO:0048555 | C | Generative cell nucleus | 1 |
| GO:0055047 | P | Generative cell mitosis | 1 |
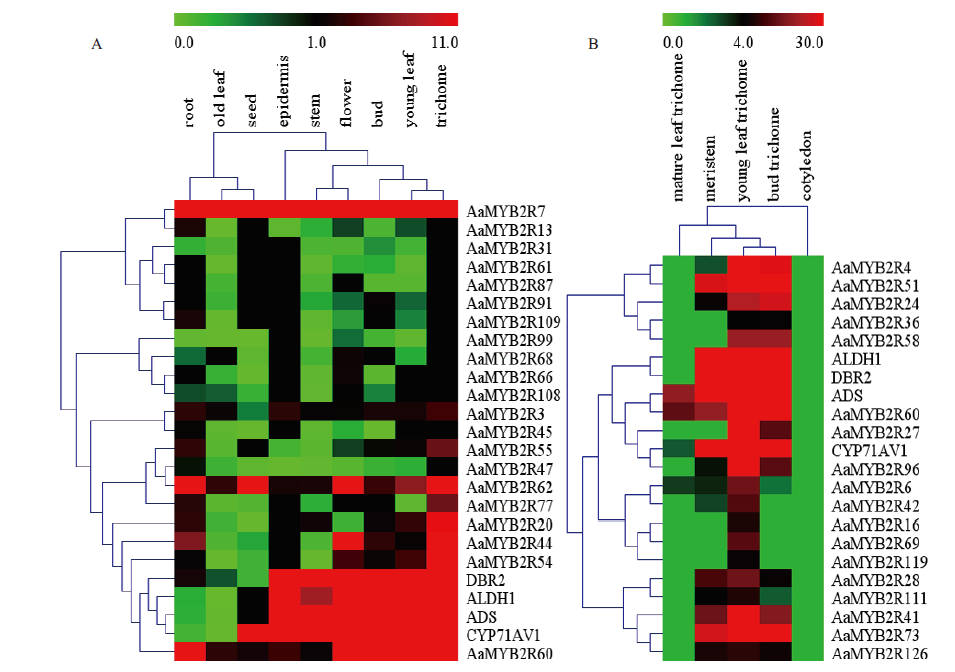
Fig.3 Expression pattern clustering of artemisinin specific synthesis pathway genes and parts of R2R3-MYB genes in A. annua A:Analysis result of expression pattern according to transcriptome data of PRJNA416223. B:Analysis result of expression pattern according to transcriptome data of PRJNA39657
| [1] | World Health Organization. World malaria report 2020[R]. Geneva:WHO, 2020. |
| [2] | Konstat-Korzenny E, Ascencio-Aragón JA, Niezen-Lugo S, et al. Artemisinin and its synthetic derivatives as a possible therapy for cancer[J]. Med Sci(Basel), 2018, 6(1):19. |
| [3] |
Mutabingwa TK. Artemisinin-based combination therapies(ACTs):best hope for malaria treatment but inaccessible to the needy![J]. Acta Trop, 2005, 95(3):305-315.
pmid: 16098946 |
| [4] |
Ro DK, Paradise EM, Ouellet M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast[J]. Nature, 2006, 440(7086):940-943.
doi: 10.1038/nature04640 URL |
| [5] | 刘德龙, 张万斌. 青蒿素可工业化合成研究进展[J]. 科学通报, 2017, 62(18):1997-2006. |
| Liu DL, Zhang WB. The development on the research of industrial production of artemisinin[J]. Chin Sci Bull, 2017, 62(18):1997-2006. | |
| [6] | 李姣, 霍晋彦, 于宗霞, 等. 生物技术在青蒿素合成中的应用[J]. 植物生理学报, 2018, 54(7):1179-1185. |
| Li J, Huo JY, Yu ZX, et al. Application of biotechnology in the biosynjournal of artemisinin[J]. Plant Physiol J, 2018, 54(7):1179-1185. | |
| [7] |
Verpoorte R, Memelink J. Engineering secondary metabolite production in plants[J]. Curr Opin Biotechnol, 2002, 13(2):181-187.
doi: 10.1016/S0958-1669(02)00308-7 URL |
| [8] | 李琦, 高晓悦, 陈万生, 等. 黄花蒿中青蒿素生物合成相关转录因子研究进展[J]. 中草药, 2021, 52(6):1827-1833. |
| Li Q, Gao XY, Chen WS, et al. Research on transcription factors related to artemisinin biosynjournal in Artemisia annua[J]. Chin Tradit Herb Drugs, 2021, 52(6):1827-1833. | |
| [9] |
Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family[J]. Plant Mol Biol, 1999, 41(5):577-585.
pmid: 10645718 |
| [10] |
Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis[J]. Trends Plant Sci, 2010, 15(10):573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [11] |
Li CN, Ng CKY, Fan LM. MYB transcription factors, active players in abiotic stress signaling[J]. Environ Exp Bot, 2015, 114:80-91.
doi: 10.1016/j.envexpbot.2014.06.014 URL |
| [12] |
Matías-Hernández L, Jiang W, Yang K, et al. AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana[J]. Plant J, 2017, 90(3):520-534.
doi: 10.1111/tpj.13509 URL |
| [13] |
Shi P, Fu X, Shen Q, et al. The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynjournal in Artemisia annua[J]. New Phytol, 2018, 217(1):261-276.
doi: 10.1111/nph.14789 pmid: 28940606 |
| [14] |
Zhou Z, Tan H, Li Q, et al. TRICHOME AND ARTEMISININ REGULATOR 2 positively regulates trichome development and artemisinin biosynjournal in Artemisia annua[J]. New Phytol, 2020, 228(3):932-945.
doi: 10.1111/nph.v228.3 URL |
| [15] |
Shen Q, Zhang L, Liao Z, et al. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynjournal[J]. Mol Plant, 2018, 11(6):776-788.
doi: 10.1016/j.molp.2018.03.015 URL |
| [16] |
Chen YH, Yang XY, He K, et al. The MYB transcription factor superfamily of Arabidopsis:expression analysis and phylogenetic comparison with the rice MYB family[J]. Plant Mol Biol, 2006, 60(1):107-124.
doi: 10.1007/s11103-005-2910-y URL |
| [17] |
Finn RD, Coggill P, Eberhardt RY, et al. The Pfam protein families database:towards a more sustainable future[J]. Nucleic Acids Res, 2016, 44(d1):D279-D285.
doi: 10.1093/nar/gkv1344 URL |
| [18] |
Li W, Godzik A. Cd-hit:a fast program for clustering and comparing large sets of protein or nucleotide sequences[J]. Bioinformatics, 2006, 22(13):1658-1659.
doi: 10.1093/bioinformatics/btl158 URL |
| [19] |
Gasteiger E, Gattiker A, Hoogland C, et al. ExPASy:The proteomics server for in-depth protein knowledge and analysis[J]. Nucleic Acids Res, 2003, 31(13):3784-3788.
doi: 10.1093/nar/gkg563 URL |
| [20] |
Crooks GE, Hon G, Chandonia JM, et al. WebLogo:a sequence logo generator[J]. Genome Res, 2004, 14(6):1188-1190.
pmid: 15173120 |
| [21] |
Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL:homology modelling of protein structures and complexes[J]. Nucleic Acids Res, 2018, 46(w1):W296-W303.
doi: 10.1093/nar/gky427 URL |
| [22] |
Bailey TL, Boden M, Buske FA, et al. MEME SUITE:tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(web server issue):W202-W208.
doi: 10.1093/nar/gkp335 URL |
| [23] |
Zheng XW, Yi DX, Shao LH, et al. In silico genome-wide identification, phylogeny and expression analysis of the R2R3-MYB gene family in Medicago truncatula[J]. J Integr Agric, 2017, 16(7):1576-1591.
doi: 10.1016/S2095-3119(16)61521-6 URL |
| [24] |
Tamura K, Stecher G, Peterson D, et al. MEGA6:molecular evolutionary genetics analysis version 6. 0[J]. Mol Biol Evol, 2013, 30(12):2725-2729.
doi: 10.1093/molbev/mst197 URL |
| [25] |
Götz S, García-Gómez JM, Terol J, et al. High-throughput functional annotation and data mining with the Blast2GO suite[J]. Nucleic Acids Res, 2008, 36(10):3420-3435.
doi: 10.1093/nar/gkn176 URL |
| [26] |
Graham I, Besser K, Blumer S, et al. The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin[J]. Science, 2010, 327(5963):328-331.
doi: 10.1126/science.1182612 pmid: 20075252 |
| [27] |
Chen C, Chen H, Zhang Y, et al. TBtools:an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 URL |
| [28] |
Patro R, Duggal G, Love MI, et al. Salmon provides fast and bias-aware quantification of transcript expression[J]. Nat Methods, 2017, 14(4):417-419.
doi: 10.1038/nmeth.4197 pmid: 28263959 |
| [29] |
Millard PS, Kragelund BB, Burow M. R2R3 MYB transcription factors-functions outside the DNA-binding domain[J]. Trends Plant Sci, 2019, 24(10):934-946.
doi: S1360-1385(19)30166-9 pmid: 31358471 |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [3] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [4] | LI Xue-qi, ZHANG Su-jie, YU Man, HUANG Jin-guang, ZHOU Huan-bin. Establishment of CRISPR/CasX-based Genome Editing Technology in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 40-48. |
| [5] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [6] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [7] | RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability [J]. Biotechnology Bulletin, 2023, 39(8): 283-290. |
| [8] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [9] | ZHANG Dao-lei, GAN Yu-jun, LE Liang, PU Li. Epigenetic Regulation of Yield-related Traits in Maize and Epibreeding [J]. Biotechnology Bulletin, 2023, 39(8): 31-42. |
| [10] | SHI Jia-xin, LIU Kai, ZHU Jin-jie, QI Xian-tao, XIE Chuan-xiao, LIU Chang-lin. Gene Editing Reshaping Maize Plant Type for Increasing Hybrid Yield [J]. Biotechnology Bulletin, 2023, 39(8): 62-69. |
| [11] | DU Dong-dong, QIAN Jing, LI Si-qi, LIU Wen-fei, WEI Xiang-li, LIU Chang-yong, LUO Rui-feng, KANG Li-chao. Whole Genome Sequencing and Analysis of Listeria monocytogenes Strain LMXJ15 [J]. Biotechnology Bulletin, 2023, 39(7): 298-306. |
| [12] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [13] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [14] | YIN Ming-hua, YU Huan-yuan, XIAO Xin-yi, WANG Yu-ting. Chloroplast Genomic Characterization and Phylogenetic Analysis of Colocasia esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan [J]. Biotechnology Bulletin, 2023, 39(6): 233-247. |
| [15] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||