








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (2): 83-94.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0664
Previous Articles Next Articles
PENG Guo-ying1( ), HU Liang1, HUANG Chao2, YANG Kun1, WAN Wei1, HUANG Chang-gan1(
), HU Liang1, HUANG Chao2, YANG Kun1, WAN Wei1, HUANG Chang-gan1( )
)
Received:2021-05-21
Online:2022-02-26
Published:2022-03-09
Contact:
HUANG Chang-gan
E-mail:1114755190@qq.com;windhcg@163.com
PENG Guo-ying, HU Liang, HUANG Chao, YANG Kun, WAN Wei, HUANG Chang-gan. Transcriptome Analysis of Response to Heavy Metal Copper Stress in Setcreasea purpurea Root Tissue[J]. Biotechnology Bulletin, 2022, 38(2): 83-94.
| 序列号Unigene ID | 注释Annotated | 序列Sequence(5'-3') | Tm/℃ | 长度Length/bp |
|---|---|---|---|---|
| c1920_g1 | 磷酸化酶Phosphorylase | F:ATGTGGGCTCTGTCGTT R:TGCCTGCTGTGCTAATG | 58/60 | 178 |
| c48219_g1 | 羧酸酯酶12 Carboxylesterase 12 | F:TTTGCGAGGTTGGGTAT R:AAGAGTGGCCGAGTTGA | 60/60 | 191 |
| c1055_g1 | 铁通透酶基因1 Fe transporter permease 1 | F:TACCTGCCCGCCGTGTT R:TGATGACTGCCGCCTCC | 60/59 | 154 |
| c1246_g1 | 金属尼古丁胺转运蛋白9 Metal nicotine amine transporter 9 | F:ATGTTAGAGTGAGATACCCTGT R:AATGGTTATGGCGTTGG | 60/60 | 177 |
| c1178_g1 | 磷酸酯酶2C Protein phosphatase 2C | F:ATTACCTACGAGGAGCG R:CCAACCTTATCCCACAG | 58/60 | 164 |
| c51710_g1 | 谷胱甘肽转移酶 Glutathione transferase | F:GAGCAAAGTAGGCATCG R:ACTAACGGCTTGAGGGT | 59/60 | 158 |
| c18811_g1 | 生长素结合蛋白 Auxin binding protein | F:GCACTGGGTCTGCCTGAT R:TGGAGTTCGCCGATGAG | 56/60 | 146 |
| c2112_g1 | 硝酸根转运蛋白 Nitrate transporter | F:CAAAGCCAACATAATACAAG R:TTGGGAGACTGAGAAGGT | 60/58 | 168 |
| c30939_g1 | 水通道蛋白 Aquaporin | F:ACCCTTCTGGCTTAGTG R:AATGATGGTGATGTGGC | 57/60 | 121 |
| c113_g1 | 假定蛋白21 Hypothetical protein 21 | F:GGCTCTTTCTGGCTCGTA R:CAAGTTCATTGGCACCC | 56/60 | 138 |
Table 1 Primer sequence of qRT-PCR
| 序列号Unigene ID | 注释Annotated | 序列Sequence(5'-3') | Tm/℃ | 长度Length/bp |
|---|---|---|---|---|
| c1920_g1 | 磷酸化酶Phosphorylase | F:ATGTGGGCTCTGTCGTT R:TGCCTGCTGTGCTAATG | 58/60 | 178 |
| c48219_g1 | 羧酸酯酶12 Carboxylesterase 12 | F:TTTGCGAGGTTGGGTAT R:AAGAGTGGCCGAGTTGA | 60/60 | 191 |
| c1055_g1 | 铁通透酶基因1 Fe transporter permease 1 | F:TACCTGCCCGCCGTGTT R:TGATGACTGCCGCCTCC | 60/59 | 154 |
| c1246_g1 | 金属尼古丁胺转运蛋白9 Metal nicotine amine transporter 9 | F:ATGTTAGAGTGAGATACCCTGT R:AATGGTTATGGCGTTGG | 60/60 | 177 |
| c1178_g1 | 磷酸酯酶2C Protein phosphatase 2C | F:ATTACCTACGAGGAGCG R:CCAACCTTATCCCACAG | 58/60 | 164 |
| c51710_g1 | 谷胱甘肽转移酶 Glutathione transferase | F:GAGCAAAGTAGGCATCG R:ACTAACGGCTTGAGGGT | 59/60 | 158 |
| c18811_g1 | 生长素结合蛋白 Auxin binding protein | F:GCACTGGGTCTGCCTGAT R:TGGAGTTCGCCGATGAG | 56/60 | 146 |
| c2112_g1 | 硝酸根转运蛋白 Nitrate transporter | F:CAAAGCCAACATAATACAAG R:TTGGGAGACTGAGAAGGT | 60/58 | 168 |
| c30939_g1 | 水通道蛋白 Aquaporin | F:ACCCTTCTGGCTTAGTG R:AATGATGGTGATGTGGC | 57/60 | 121 |
| c113_g1 | 假定蛋白21 Hypothetical protein 21 | F:GGCTCTTTCTGGCTCGTA R:CAAGTTCATTGGCACCC | 56/60 | 138 |
| 样本 Sample | 原始数据 Raw reads/Mb | 过滤数据 Clean reads /Mb | 碱基总数 Total number of bases/Gb | Q20/% | Q30/% | 过滤数据率 Clean reads ratio/% |
|---|---|---|---|---|---|---|
| CK-1 | 46.39 | 42.94 | 7.00 | 96.91 | 92.50 | 92.58 |
| CK-2 | 44.77 | 41.86 | 6.76 | 96.86 | 92.44 | 93.51 |
| CK-3 | 43.28 | 40.72 | 6.54 | 96.88 | 92.36 | 94.07 |
| CT1-1 | 44.58 | 41.71 | 6.73 | 96.82 | 92.34 | 93.56 |
| CT1-2 | 50.21 | 47.17 | 7.58 | 96.89 | 92.36 | 93.95 |
| CT1-3 | 44.60 | 41.94 | 6.73 | 96.67 | 91.91 | 94.03 |
| CT2-1 | 47.85 | 44.95 | 7.23 | 96.79 | 92.17 | 93.93 |
| CT2-2 | 40.67 | 38.21 | 6.14 | 96.92 | 92.43 | 93.95 |
| CT2-3 | 45.70 | 42.69 | 6.90 | 96.91 | 92.42 | 93.40 |
Table 2 Data statistics of disembarkation
| 样本 Sample | 原始数据 Raw reads/Mb | 过滤数据 Clean reads /Mb | 碱基总数 Total number of bases/Gb | Q20/% | Q30/% | 过滤数据率 Clean reads ratio/% |
|---|---|---|---|---|---|---|
| CK-1 | 46.39 | 42.94 | 7.00 | 96.91 | 92.50 | 92.58 |
| CK-2 | 44.77 | 41.86 | 6.76 | 96.86 | 92.44 | 93.51 |
| CK-3 | 43.28 | 40.72 | 6.54 | 96.88 | 92.36 | 94.07 |
| CT1-1 | 44.58 | 41.71 | 6.73 | 96.82 | 92.34 | 93.56 |
| CT1-2 | 50.21 | 47.17 | 7.58 | 96.89 | 92.36 | 93.95 |
| CT1-3 | 44.60 | 41.94 | 6.73 | 96.67 | 91.91 | 94.03 |
| CT2-1 | 47.85 | 44.95 | 7.23 | 96.79 | 92.17 | 93.93 |
| CT2-2 | 40.67 | 38.21 | 6.14 | 96.92 | 92.43 | 93.95 |
| CT2-3 | 45.70 | 42.69 | 6.90 | 96.91 | 92.42 | 93.40 |
| 数据库 Database | 注释基因的数量Number of annotated unigenes | 注释基因的比例Percentage of annotated unigenes/% |
|---|---|---|
| NR | 68 227 | 82.73 |
| GO | 23 236 | 28.17 |
| KEGG | 35 123 | 42.59 |
| Pfam | 27 806 | 33.72 |
| eggNOG | 67 602 | 81.97 |
| Swissprot | 52 994 | 64.26 |
| In all database | 8 422 | 10.21 |
Table 3 Summary of database annotation results
| 数据库 Database | 注释基因的数量Number of annotated unigenes | 注释基因的比例Percentage of annotated unigenes/% |
|---|---|---|
| NR | 68 227 | 82.73 |
| GO | 23 236 | 28.17 |
| KEGG | 35 123 | 42.59 |
| Pfam | 27 806 | 33.72 |
| eggNOG | 67 602 | 81.97 |
| Swissprot | 52 994 | 64.26 |
| In all database | 8 422 | 10.21 |
| 对照组Control | 实验组Case | 上调基因数Number of up-regulated gene | 下调基因数Number of down-regulated gene | 差异表达基因总数Total DEGs |
|---|---|---|---|---|
| CK | CT1 | 3 138 | 1 890 | 5 028 |
| CK | CT2 | 5 061 | 4 921 | 9 982 |
| CT1 | CT2 | 2 555 | 4 258 | 6 813 |
Table 4 Statistical analysis of differentially expressed genes in S. purpurea roots
| 对照组Control | 实验组Case | 上调基因数Number of up-regulated gene | 下调基因数Number of down-regulated gene | 差异表达基因总数Total DEGs |
|---|---|---|---|---|
| CK | CT1 | 3 138 | 1 890 | 5 028 |
| CK | CT2 | 5 061 | 4 921 | 9 982 |
| CT1 | CT2 | 2 555 | 4 258 | 6 813 |
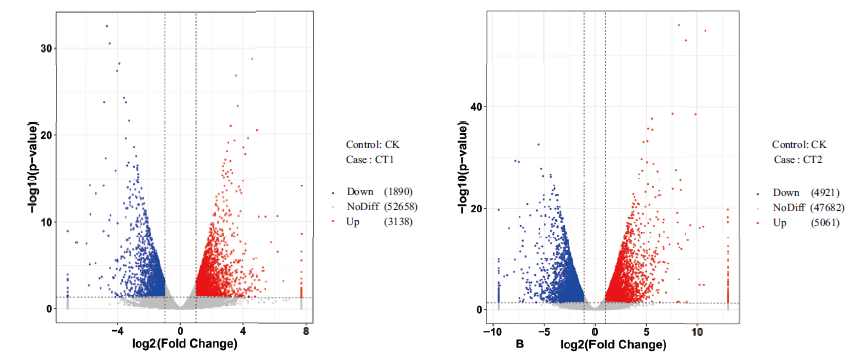
Fig. 2 Volcano map of differentially expressed genes The blue dot region on the left refers to significantly down-regulated genes,the red dot region on the right refers to significantly up-regulated genes,and the gray dot region on the bottom refers to non-significantly differentially expressed gene
| KEGG ID | 注释 Annotated | 上调基因数 Number of up-regulated genes | 下调基因数Number of down-regulated genes | 差异表达基因总数 Total DEGs | P值 P value | |
|---|---|---|---|---|---|---|
| CT1 vs CK | ko00940 | 类苯基丙酸合成 Phenylpropanoid biosynthesis | 78 | 21 | 99 | 1.902E-30 |
| ko00500 | 淀粉和蔗糖代谢 Starch and sucrose metabolism | 54 | 30 | 84 | 1.265E-15 | |
| ko04075 | 植物激素信号转导 Plant hormone signal transduction | 31 | 1 | 32 | 1.298E-13 | |
| CT2 vs CT1 | ko04016 | 植物MAPK信号通路 Plant MAPK signaling pathway | 89 | 84 | 173 | 3.056E-26 |
| ko04075 | 植物激素信号转导 Plant hormone signal transduction | 62 | 82 | 144 | 5.213E-23 | |
| ko04146 | 过氧物酶体 Peroxisome | 65 | 106 | 171 | 2.283E-17 |
Table 5 The significant KEGG enrichment pathway in S. purpurea transcriptome
| KEGG ID | 注释 Annotated | 上调基因数 Number of up-regulated genes | 下调基因数Number of down-regulated genes | 差异表达基因总数 Total DEGs | P值 P value | |
|---|---|---|---|---|---|---|
| CT1 vs CK | ko00940 | 类苯基丙酸合成 Phenylpropanoid biosynthesis | 78 | 21 | 99 | 1.902E-30 |
| ko00500 | 淀粉和蔗糖代谢 Starch and sucrose metabolism | 54 | 30 | 84 | 1.265E-15 | |
| ko04075 | 植物激素信号转导 Plant hormone signal transduction | 31 | 1 | 32 | 1.298E-13 | |
| CT2 vs CT1 | ko04016 | 植物MAPK信号通路 Plant MAPK signaling pathway | 89 | 84 | 173 | 3.056E-26 |
| ko04075 | 植物激素信号转导 Plant hormone signal transduction | 62 | 82 | 144 | 5.213E-23 | |
| ko04146 | 过氧物酶体 Peroxisome | 65 | 106 | 171 | 2.283E-17 |
| 基因序列号Unigene ID | 注释 Annotated | RNA-seq | |||||
|---|---|---|---|---|---|---|---|
| Log2FC(CT1/CK) | P值P-value | Log2FC(CT2/CK) | P值P value | ||||
| c76816_g1 | 重金属转运ATP酶5 Heavy metal transporting ATPase 5,HMA5 | 1.61 | 3.122E-05 | 0.97 | 1.502E-06 | ||
| c34452_g1 | 谷胱甘肽过氧化物酶Glutathione peroxidase,GPX | 2.95 | 1.521E-08 | 4.12 | 3.830E-07 | ||
| c14543_g1 | 铜锌超氧化物歧化酶 Cu/Zn superoxide dismutase,Cu/Zn-SOD | 1.22 | 0.000442 | 1.08 | 0.000131 | ||
| c11849_g1 | 金属硫蛋白Metallothioneins,MTs | 1.67 | 0.000924 | 3.37 | 0.000210 | ||
| c44409_g1 | 植物络合素合酶Phytochelatin synthase,PCS | 0.49 | 0.000148 | 1.21 | 0.000251 | ||
Table 6 Analysis of genes related to copper stress
| 基因序列号Unigene ID | 注释 Annotated | RNA-seq | |||||
|---|---|---|---|---|---|---|---|
| Log2FC(CT1/CK) | P值P-value | Log2FC(CT2/CK) | P值P value | ||||
| c76816_g1 | 重金属转运ATP酶5 Heavy metal transporting ATPase 5,HMA5 | 1.61 | 3.122E-05 | 0.97 | 1.502E-06 | ||
| c34452_g1 | 谷胱甘肽过氧化物酶Glutathione peroxidase,GPX | 2.95 | 1.521E-08 | 4.12 | 3.830E-07 | ||
| c14543_g1 | 铜锌超氧化物歧化酶 Cu/Zn superoxide dismutase,Cu/Zn-SOD | 1.22 | 0.000442 | 1.08 | 0.000131 | ||
| c11849_g1 | 金属硫蛋白Metallothioneins,MTs | 1.67 | 0.000924 | 3.37 | 0.000210 | ||
| c44409_g1 | 植物络合素合酶Phytochelatin synthase,PCS | 0.49 | 0.000148 | 1.21 | 0.000251 | ||
| [1] |
Watanabe ME. Phytoremediation on the brink of commericialization[J]. Environ Sci Technol, 1997, 31(4):182A-186A.
doi: 10.1021/es972219s URL |
| [2] |
Naila A, Meerdink G, Jayasena V, et al. A review on global metal accumulators-mechanism, enhancement, commercial application, and research trend[J]. Environ Sci Pollut Res Int, 2019, 26(26):26449-26471.
doi: 10.1007/s11356-019-05992-4 URL |
| [3] |
Qiao XQ, Zheng ZZ, Zhang LF, et al. Lead tolerance mechanism in sterilized seedlings of Potamogeton crispus L. :Subcellular distribution, polyamines and proline[J]. Chemosphere, 2015, 120:179-187.
doi: 10.1016/j.chemosphere.2014.06.055 URL |
| [4] |
Benyó D, Horváth E, Németh E, et al. Physiological and molecular responses to heavy metal stresses suggest different detoxification mechanism of Populus deltoides and P. x canadensis[J]. J Plant Physiol, 2016, 201:62-70.
doi: 10.1016/j.jplph.2016.05.025 URL |
| [5] |
Li Y, Zhou C, et al. Lead tolerance mechanism in Conyza canad-ensis:subcellular distribution, ultrastructure, antioxidative defense system, and phytochelatins[J]. J Plant Res, 2016, 129(2):251-262.
doi: 10.1007/s10265-015-0776-x URL |
| [6] |
Thakur S, Singh L, Wahid ZA, et al. Plant-driven removal of heavy metals from soil:uptake, translocation, tolerance mechanism, challenges, and future perspectives[J]. Environ Monit Assess, 2016, 188(4):1-11.
doi: 10.1007/s10661-015-4999-z URL |
| [7] |
Chen S, Wang Q, Lu HL, et al. Phenolic metabolism and related heavy metal tolerance mechanism in Kandelia Obovata under Cd and Zn stress[J]. Ecotoxicol Environ Saf, 2019, 169:134-143.
doi: 10.1016/j.ecoenv.2018.11.004 URL |
| [8] | 黄长干. 紫鸭跖草对铜的积累规律及在铜胁迫下的生理反应研究[D]. 长沙:湖南农业大学, 2007. |
| Huang CG. Study on copper accumulation rule and physiological responses to copper stress in setcreasea purpurea boom[D]. Changsha:Hunan Agricultural University, 2007. | |
| [9] | 杨坤, 黄超, 卢山, 等. 铜胁迫下紫鸭跖草根组织实时定量PCR内参基因的选择[J]. 植物生理学报, 2021, 57(1):195-204. |
| Yang K, Huang C, Lu S, et al. Reference gene selection for quantitative real-time PCR in purple setcreasea(Setcreasea purpurea)root tissue under copper stress[J]. Plant Physiol J, 2021, 57(1):195-204. | |
| [10] |
Labbé S, Thiele DJ. Pipes and wiring:the regulation of copper uptake and distribution in yeast[J]. Trends Microbiol, 1999, 7(12):500-505.
pmid: 10603486 |
| [11] | 周博如, 王雷, 等. 转金属硫蛋白基因(MT1)烟草耐NaCl胁迫能力[J]. 生态学报, 2010, 30(15):4103-4108. |
| Zhou BR, Wang L, et al. Characterization of transgenic tobacco overexpressing metallothionein gene(MT1)on NaCl stress[J]. Acta Ecol Sin, 2010, 30(15):4103-4108. | |
| [12] |
Cobbett C, Goldsbrough P. Phytochelatins and metallothioneins:roles in heavy metal detoxification and homeostasis[J]. Annu Rev Plant Biol, 2002, 53:159-182.
doi: 10.1146/arplant.2002.53.issue-1 URL |
| [13] | 林芳竹. 叶用红菾菜重金属转运蛋白基因克隆研究[D]. 沈阳:沈阳大学, 2019. |
| Lin FZ. Gene cloning in heavy metal transport protein of beta vulgars var. cicla L[D]. Shenyang:Shenyang University, 2019. | |
| [14] | 李洁. 苦荞ABC转运蛋白基因的克隆与功能初步分析[D]. 杨凌:西北农林科技大学, 2018. |
| Li J. Cloning and function analysis of ABC transporter gene from Tartary buckwheat[D]. Yangling:Northwest A & F University, 2018. | |
| [15] | 钟茜. 水稻P1B型ATPase重金属转运蛋白的结构与功能研究进展[J]. 宁夏师范学院学报, 2013, 34(6):62-69. |
| Zhong Q. Structure and function of heavy metal transporter P1B-ATPase in rice:a Review[J]. J Ningxia Teach Univ, 2013, 34(6):62-69. | |
| [16] |
Colangelo EP, Guerinot ML. Put the metal to the petal:metal uptake and transport throughout plants[J]. Curr Opin Plant Biol, 2006, 9(3):322-330.
doi: 10.1016/j.pbi.2006.03.015 URL |
| [17] | 王聪. 橡胶树铜离子转运蛋白HbCOPT基因克隆与功能验证[D]. 海口:海南大学, 2017. |
| Wang C. Cloning and function identification of copper transporter genes COPTs from Hevea brasiliensis[D]. Haikou:Hainan University, 2017. | |
| [18] |
刘元峰, 李素贞, 郭晋杰, 等. 植物YSL家族基因研究进展[J]. 生物技术通报, 2017, 33(9):1-9.
doi: 10.13560/j.cnki.biotech.bull.1985.2017-0375 |
| Liu YF, Li SZ, Guo JJ, et al. Research progress on YSL transporters gene family[J]. Biotechnol Bull, 2017, 33(9):1-9. | |
| [19] | Li XM, Gan P, Liang Y, et al. Specific protein properties of setcreasea pupurea boom under copper stress[J]. Agric Sci Technol, 2012, 13(5):942-944. |
| [20] | 王钦仪, 李斯濛, 郁培义, 等 铅锌胁迫下山苍子雌雄花蕾转录组测序分析[J]. 分子植物育种, 2021. https://kns.cnki.net/kcms/detail/46.1068.s.20210412.1446.010.html . |
| Wang QY, Li SM, Yu PY, et al. Analysis of Pb and Zn stress on transcriptome sequencing of male and female flower buds in Litsea cubeba(Lour. )Pers[J]. Molecular Plant Breeding, 2021. https://kns.cnki.net/kcms/detail/46.1068.s.20210412.1446.010.html . | |
| [21] | 刘佳, 田云, 邓家林. 碱胁迫下榆叶梅差异基因表达分析[J]. 核农学报, 2020, 34(7):1397-1408. |
| Liu J, Tian Y, Deng JL. Differential gene expression analysis of Prunus triloba lindl. under alkaline stress[J]. J Nucl Agric Sci, 2020, 34(7):1397-1408. | |
| [22] | 韦晓霞, 王小安, 等. 百香果低温胁迫转录组及茉莉酸代谢基因分析[J]. 核农学报, 2021, 35(4):815-825. |
| Wei XX, Wang XA, et al. Transcriptome and jasmin metabolism gene analysis of Passiflora edulia Sims under low temperature stress[J]. J Nucl Agric Sci, 2021, 35(4):815-825. | |
| [23] |
Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool[J]. J Mol Biol, 1990, 215(3):403-410.
doi: 10.1016/S0022-2836(05)80360-2 pmid: 2231712 |
| [24] |
Liu FG, Tang XQ, Sheng XZ, et al. Edwardsiella tarda outer membrane protein C:an immunogenic protein induces highly protective effects in flounder(Paralichthys olivaceus)against edwardsiellosis[J]. Int J Mol Sci, 2016, 17(7):1117.
doi: 10.3390/ijms17071117 URL |
| [25] |
Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data[J]. Nat Methods, 2010, 7(5):335-336.
doi: 10.1038/nmeth.f.303 pmid: 20383131 |
| [26] |
Hawkins RD, Hon GC. Next-generation genomics:an integrative approach[J]. Nat Rev Genet, 2010, 11(7):476-486.
doi: 10.1038/nrg2795 pmid: 20531367 |
| [27] |
Annadurai RS, Jayakumar V, Mugasimangalam RC, et al. Next generation sequencing and de novo transcriptome analysis of Costus pictus D. Don, a non-model plant with potent anti-diabetic properties[J]. BMC Genomics, 2012, 13:663.
doi: 10.1186/1471-2164-13-663 pmid: 23176672 |
| [28] |
Türktaş M, Yücebİlgİlİ Kurtoğlu K, et al. Sequencing of plant genomes - a review[J]. Turk J Agric For, 2015, 39:361-376.
doi: 10.3906/tar-1409-93 URL |
| [29] | Daniel K, José MJG, Seisuke K, et al. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato[J]. PNAS. 2013, 110(28):E2655-2662. |
| [30] |
Li Y, Huang J, Song XW, et al. An RNA-Seq transcriptome analysis revealing novel insights into aluminum tolerance and accumulation in tea plant[J]. Planta, 2017, 246(1):91-103.
doi: 10.1007/s00425-017-2688-6 URL |
| [31] |
Rathi D, Gayali S, et al. Transcriptome profiling illustrates expression signatures of dehydration tolerance in developing grasspea seedlings[J]. Planta, 2019, 250(3):839-855.
doi: 10.1007/s00425-018-03082-2 URL |
| [32] |
Taheri S, Lee Abdullah T, Yusop M, et al. Mining and development of novel SSR markers using next generation sequencing(NGS)data in plants[J]. Molecules, 2018, 23(2):399.
doi: 10.3390/molecules23020399 URL |
| [33] |
Rai A, Nakamura M, Takahashi H, et al. High-throughput sequencing and de novo transcriptome assembly of Swertia japonica to identify genes involved in the biosynjournal of therapeutic metabolites[J]. Plant Cell Rep, 2016, 35(10):2091-2111.
doi: 10.1007/s00299-016-2021-z URL |
| [34] |
Andrés-Colás N, Sancenón V, Rodríguez-Navarro S, et al. The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots[J]. Plant J, 2006, 45(2):225-236.
pmid: 16367966 |
| [35] | Wang K, Tang SF, Hou XM. Molecular mechanism investigation on the interactions of copper(II)ions with glutathione peroxidase 6 from Arabidopsis thaliana[J]. Spectrochimica Acta A:Mol Biomol Spectrosc, 2018, 203:428-433. |
| [36] |
Wang W, Xia MX, Chen J, et al. Gene expression characteristics and regulation mechanisms of superoxide dismutase and its physiological roles in plants under stress[J]. Biochemistry:Mosc, 2016, 81(5):465-480.
doi: 10.1134/S0006297916050047 URL |
| [37] |
Zhang HX, Lv S, Xu HW, et al. H2O2 is involved in the metallothionein-mediated rice tolerance to copper and cadmium toxicity[J]. Int J Mol Sci, 2017, 18(10):2083.
doi: 10.3390/ijms18102083 URL |
| [38] | 李慧, 丛郁, 王宏伟, 等. 豆梨植物络合素合酶PcPCS1基因克隆及其表达分析[J]. 园艺学报, 2010, 37(6):880-890. |
| Li H, Cong Y, Wang HW, et al. Molecular cloning and expression analysis of a phytochelatin synthase gene, PcPCS1, from Pyrus calleryana dcne[J]. Acta Hortic Sin, 2010, 37(6):880-890. |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [3] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [4] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [5] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [6] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| [7] | HU Li-li, LIN Bo-rong, WANG Hong-hong, CHEN Jian-song, LIAO Jin-ling, ZHUO Kan. Transcriptome and Candidate Effectors Analysis of Pratylenchus brachyurus [J]. Biotechnology Bulletin, 2023, 39(3): 254-266. |
| [8] | SUN Yan-qiu, XIE Cai-yun, TANG Yue-qin. Construction and Mechanism Analysis of High-temperature Resistant Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(11): 226-237. |
| [9] | XU Jun, YE Yu-qing, NIU Ya-jing, HUANG He, ZHANG Meng-meng. Transcriptome Analysis of Rhizome Development in Chrysanthemum× × morifolium [J]. Biotechnology Bulletin, 2023, 39(10): 231-245. |
| [10] | LUO Hao-tian, WANG Long, WANG Yu-qian, WANG Yue, LI Jia-zhen, YANG Meng-ke, ZHANG Jie, DENG Xin, WANG Hong-yan. Genome-wide Identification and Expression Analysis of the RNAi-related Gene Families in Setaria viridis [J]. Biotechnology Bulletin, 2023, 39(1): 175-186. |
| [11] | XIN Jian-pan, LI Yan, ZHAO Chu, TIAN Ru-nan. Transcriptome Sequencing in the Leaves of Pontederia cordata with Cadmium Exposure and Gene Mining in Phenypropanoid Pathways [J]. Biotechnology Bulletin, 2022, 38(6): 198-210. |
| [12] | XU Jin, LI Tao, LI Chu-lin, ZHU Shun-ni, WANG Zhong-ming, XIANG Wen-zhou. Effects of Temperature on the Growth,Total Lipid and Eicosapentaenoic Acid Synthesis of Eustigmatos sp. [J]. Biotechnology Bulletin, 2022, 38(6): 261-271. |
| [13] | XIONG He-li, SHA Qian, LIU Shao-na, XIANG De-cai, ZHANG Bin, ZHAO Zhi-yong. Application of Single-cell Transcriptome Sequencing in Animals [J]. Biotechnology Bulletin, 2022, 38(3): 226-233. |
| [14] | ZHANG Bin, YANG Xin-xia. Identification of Key Transcription Factors in Response to Salt Stress in Rice [J]. Biotechnology Bulletin, 2022, 38(3): 9-15. |
| [15] | GUAN Yi, LI Xin, WANG Ding-yi, DU Xi, ZHANG Long-bin, YE Xiu-yun. Functional Study of BbRho5 on the Growth Rate of Beauveria bassiana [J]. Biotechnology Bulletin, 2022, 38(2): 132-140. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||