








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (9): 77-85.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0717
Previous Articles Next Articles
WANG Qi( ), WU Zhi-xuan, CHEN Zhong-ling, WU Bai-yi-la, HU Zong-fu(
), WU Zhi-xuan, CHEN Zhong-ling, WU Bai-yi-la, HU Zong-fu( ), NIU Hua-xin(
), NIU Hua-xin( )
)
Received:2021-06-07
Online:2021-09-26
Published:2021-10-25
Contact:
HU Zong-fu,NIU Hua-xin
E-mail:wangqi@163.com;huzongfusohu@163.com;niuhx@imun.edu.cn
WANG Qi, WU Zhi-xuan, CHEN Zhong-ling, WU Bai-yi-la, HU Zong-fu, NIU Hua-xin. Effects of Lactobacillus paracasei on the Quality and Bacterial Diversity of Silage Alfalfa After Aerobic Exposure[J]. Biotechnology Bulletin, 2021, 37(9): 77-85.
| 组别Group | 鲜样Fresh sample | PO0 | PO7 | PO14 | SEM | ANOVA | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CON | LCP | CON | LCP | CON | LCP | I | S | I×S | |||
| DM /% | 23.92 | 20.85 | 22.26 | 20.57 | 21.49 | 20.79 | 21.72 | 0.48 | ** | NS | NS |
| pH | 6.27 | 5.46 | 4.55 | 5.44 | 4.74 | 6.43 | 4.83 | 0.01 | ** | ** | ** |
| LAB/(log10 CFU/g FM) | 5.53 | 7.22 | 7.68 | 6.56 | 7.34 | 5.38 | 6.28 | 0.06 | ** | ** | ** |
| Yeasts/(log10 CFU/g FM) | 3.76 | 3.28 | 2.56 | 4.46 | 3.90 | 7.13 | 6.07 | 0.09 | ** | ** | ** |
| Molds/(log10 CFU/g FM) | 3.92 | 2.80 | <2.00 | 3.08 | 3.51 | 6.24 | 4.68 | 0.09 | — | — | — |
| LA/(g·kg-1 DM) | 31.67 | 48.61 | 11.56 | 26.85 | 4.73 | 15.49 | 1.04 | ** | ** | ** | |
| AA/(g·kg-1 DM) | 18.32 | 15.88 | 11.55 | 12.52 | 7.43 | 10.41 | 0.63 | ** | ** | ** | |
| PA/(g·kg-1 DM) | 3.75 | 2.64 | 1.56 | 1.24 | 0.82 | 0.68 | 0.28 | * | ** | NS | |
| BA/(g·kg-1 DM) | 1.43 | 0.45 | 2.01 | 1.48 | 3.21 | 2.61 | 0.04 | ** | ** | ** | |
Table 1 Fermentation quality of alfalfa silage during aerobic exposure from 0 to 14 d
| 组别Group | 鲜样Fresh sample | PO0 | PO7 | PO14 | SEM | ANOVA | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CON | LCP | CON | LCP | CON | LCP | I | S | I×S | |||
| DM /% | 23.92 | 20.85 | 22.26 | 20.57 | 21.49 | 20.79 | 21.72 | 0.48 | ** | NS | NS |
| pH | 6.27 | 5.46 | 4.55 | 5.44 | 4.74 | 6.43 | 4.83 | 0.01 | ** | ** | ** |
| LAB/(log10 CFU/g FM) | 5.53 | 7.22 | 7.68 | 6.56 | 7.34 | 5.38 | 6.28 | 0.06 | ** | ** | ** |
| Yeasts/(log10 CFU/g FM) | 3.76 | 3.28 | 2.56 | 4.46 | 3.90 | 7.13 | 6.07 | 0.09 | ** | ** | ** |
| Molds/(log10 CFU/g FM) | 3.92 | 2.80 | <2.00 | 3.08 | 3.51 | 6.24 | 4.68 | 0.09 | — | — | — |
| LA/(g·kg-1 DM) | 31.67 | 48.61 | 11.56 | 26.85 | 4.73 | 15.49 | 1.04 | ** | ** | ** | |
| AA/(g·kg-1 DM) | 18.32 | 15.88 | 11.55 | 12.52 | 7.43 | 10.41 | 0.63 | ** | ** | ** | |
| PA/(g·kg-1 DM) | 3.75 | 2.64 | 1.56 | 1.24 | 0.82 | 0.68 | 0.28 | * | ** | NS | |
| BA/(g·kg-1 DM) | 1.43 | 0.45 | 2.01 | 1.48 | 3.21 | 2.61 | 0.04 | ** | ** | ** | |
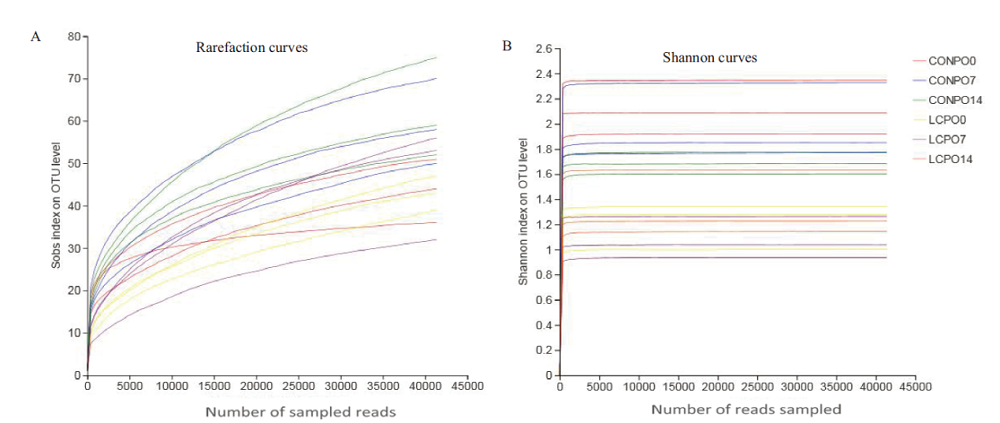
Fig.1 Rarefaction curves(A)and Shannon curves(B)of bacterial community in alfalfa silage CONPO0,CONPO7 and CONPO14 represent the control group aerobic exposure 0,7 and 14 d,respectively. LCPPO0,LCPPO7 and LCPPO14 represent the Lactobacillus casei inoculation treatment group aerobic exposure 0,7 and 14 d,respectively
| Sample\Estimators | Sobs | Shannon | Simpson | Ace | Chao |
|---|---|---|---|---|---|
| CONPO0_1 | 43.67 | 2.12 | 0.18 | 53.25 | 49.37 |
| CONPO7_1 | 59.33 | 1.98 | 0.22 | 71.99 | 67.07 |
| CONPO14_1 | 62.00 | 1.69 | 0.33 | 79.05 | 74.23 |
| LCPPO0_1 | 43.00 | 1.21 | 0.41 | 79.80 | 55.789 |
| LCPPO7_1 | 47.00 | 1.08 | 0.49 | 76.73 | 60.91 |
| LCPPO14_1 | 44.00 | 1.33 | 0.38 | 92.71 | 67.73 |
| SEM | 2.66 | 0.10 | 0.03 | 5.65 | 3.22 |
| P | 0.092 | 0.001 | 0.004 | 0.552 | 0.263 |
Table 2 Alpha-diversity of bacterial community in the silage alfalfa during aerobic exposure
| Sample\Estimators | Sobs | Shannon | Simpson | Ace | Chao |
|---|---|---|---|---|---|
| CONPO0_1 | 43.67 | 2.12 | 0.18 | 53.25 | 49.37 |
| CONPO7_1 | 59.33 | 1.98 | 0.22 | 71.99 | 67.07 |
| CONPO14_1 | 62.00 | 1.69 | 0.33 | 79.05 | 74.23 |
| LCPPO0_1 | 43.00 | 1.21 | 0.41 | 79.80 | 55.789 |
| LCPPO7_1 | 47.00 | 1.08 | 0.49 | 76.73 | 60.91 |
| LCPPO14_1 | 44.00 | 1.33 | 0.38 | 92.71 | 67.73 |
| SEM | 2.66 | 0.10 | 0.03 | 5.65 | 3.22 |
| P | 0.092 | 0.001 | 0.004 | 0.552 | 0.263 |
| Genus | OTU | CONPO0 | CONPO7 | CONPO14 | LCPPO0 | LCPPO7 | LCPPO14 |
|---|---|---|---|---|---|---|---|
| Acetobacter | OTU53 | 0 | 0.03 | 34.99 | 0 | 0.27 | 18.37 |
| Lactobacillus | OTU123 | 0 | 0 | 0 | 4.08 | 0.35 | 0.46 |
| Pediococcus | OTU58 | 0 | 0 | 0 | 0.52 | 1.2 | 0.13 |
| Weissella | OTU26 | 4.17 | 3.71 | 1.58 | 0 | 0.01 | 0 |
| Komagataeibacter | OTU125 | 0 | 0 | 0 | 0 | 5.58 | |
| Cedecea | OTU45 | 2.02 | 1.81 | 3.29 | 0 | 0 | 0 |
| Lactobacillus | OTU127 | 0.65 | 0.48 | 0.19 | 0 | 0.01 | 2.29 |
| Lactobacillus | OTU133 | 0.64 | 1.7 | 0.84 | 9.2 | 17.81 | 21.23 |
| Lactobacillus | OTU41 | 2.63 | 34.63 | 28.38 | 0.18 | 3.97 | |
| Lysinibacillus | OTU57 | 0 | 0 | 3.81 | 0 | 0 | 0 |
| ALCPaligenes | OTU35 | 0 | 0 | 0.36 | 0 | 0 | 0.03 |
| Sporolactobacillus | OTU29 | 1.1 | 0.12 | 0.03 | 0 | 0 | 0 |
| Lactobacillus | OTU27 | 8.52 | 10.71 | 8.47 | 0.06 | 0.05 | 0.13 |
| Lactobacillus | OTU153 | 0.32 | 0.05 | 52.99 | 65.39 | 42.11 | |
| Garciella | OTU11 | 7.74 | 0 | 0 | 0 | 0 | 0 |
| Lactobacillus | OTU54 | 4.3 | 3.17 | 2.54 | 0 | 0 | 0 |
| Enterobacter | OTU38 | 32.58 | 19.35 | 6.59 | 0.94 | 1.19 | 0.43 |
| Enterococcus | OTU21 | 13.12 | 6.84 | 3.19 | 0.04 | 0.03 | 0.03 |
| Lactobacillus | OTU104 | 3.74 | 1.19 | 0.27 | 26.65 | 9.07 | 3.81 |
| Lactobacillus | OTU76 | 1.23 | 8.69 | 1.47 | 0.07 | 0.05 | 0.06 |
| Lactobacillus | OTU148 | 0 | 0 | 0 | 1.98 | 1.74 | 0.2 |
| Lactobacillus | OTU115 | 0.01 | 0.01 | 0 | 2.54 | 1.69 | 0.85 |
| Lactobacillus | OTU71 | 2.95 | 4.36 | 1.68 | 0 | 0 | 0 |
| Enterococcus | OTU14 | 3.79 | 1.34 | 0.67 | 0 | 0 | 0 |
Table 3 OTU composition of the silage alfalfa in different exposure time and treatments(>1%)
| Genus | OTU | CONPO0 | CONPO7 | CONPO14 | LCPPO0 | LCPPO7 | LCPPO14 |
|---|---|---|---|---|---|---|---|
| Acetobacter | OTU53 | 0 | 0.03 | 34.99 | 0 | 0.27 | 18.37 |
| Lactobacillus | OTU123 | 0 | 0 | 0 | 4.08 | 0.35 | 0.46 |
| Pediococcus | OTU58 | 0 | 0 | 0 | 0.52 | 1.2 | 0.13 |
| Weissella | OTU26 | 4.17 | 3.71 | 1.58 | 0 | 0.01 | 0 |
| Komagataeibacter | OTU125 | 0 | 0 | 0 | 0 | 5.58 | |
| Cedecea | OTU45 | 2.02 | 1.81 | 3.29 | 0 | 0 | 0 |
| Lactobacillus | OTU127 | 0.65 | 0.48 | 0.19 | 0 | 0.01 | 2.29 |
| Lactobacillus | OTU133 | 0.64 | 1.7 | 0.84 | 9.2 | 17.81 | 21.23 |
| Lactobacillus | OTU41 | 2.63 | 34.63 | 28.38 | 0.18 | 3.97 | |
| Lysinibacillus | OTU57 | 0 | 0 | 3.81 | 0 | 0 | 0 |
| ALCPaligenes | OTU35 | 0 | 0 | 0.36 | 0 | 0 | 0.03 |
| Sporolactobacillus | OTU29 | 1.1 | 0.12 | 0.03 | 0 | 0 | 0 |
| Lactobacillus | OTU27 | 8.52 | 10.71 | 8.47 | 0.06 | 0.05 | 0.13 |
| Lactobacillus | OTU153 | 0.32 | 0.05 | 52.99 | 65.39 | 42.11 | |
| Garciella | OTU11 | 7.74 | 0 | 0 | 0 | 0 | 0 |
| Lactobacillus | OTU54 | 4.3 | 3.17 | 2.54 | 0 | 0 | 0 |
| Enterobacter | OTU38 | 32.58 | 19.35 | 6.59 | 0.94 | 1.19 | 0.43 |
| Enterococcus | OTU21 | 13.12 | 6.84 | 3.19 | 0.04 | 0.03 | 0.03 |
| Lactobacillus | OTU104 | 3.74 | 1.19 | 0.27 | 26.65 | 9.07 | 3.81 |
| Lactobacillus | OTU76 | 1.23 | 8.69 | 1.47 | 0.07 | 0.05 | 0.06 |
| Lactobacillus | OTU148 | 0 | 0 | 0 | 1.98 | 1.74 | 0.2 |
| Lactobacillus | OTU115 | 0.01 | 0.01 | 0 | 2.54 | 1.69 | 0.85 |
| Lactobacillus | OTU71 | 2.95 | 4.36 | 1.68 | 0 | 0 | 0 |
| Enterococcus | OTU14 | 3.79 | 1.34 | 0.67 | 0 | 0 | 0 |

Fig. 7 LEfSe analysis of the bacteria taxon with significant difference between treatments(LAD>4.0) Nodes with different colors indicate microbial groups that are significantly enriched in the corresponding groups and have a significant impact on the differences between groups. Light yellow nodes indicate microbial groups that have no significant differences in different groups or have no significant impact on differences between groups
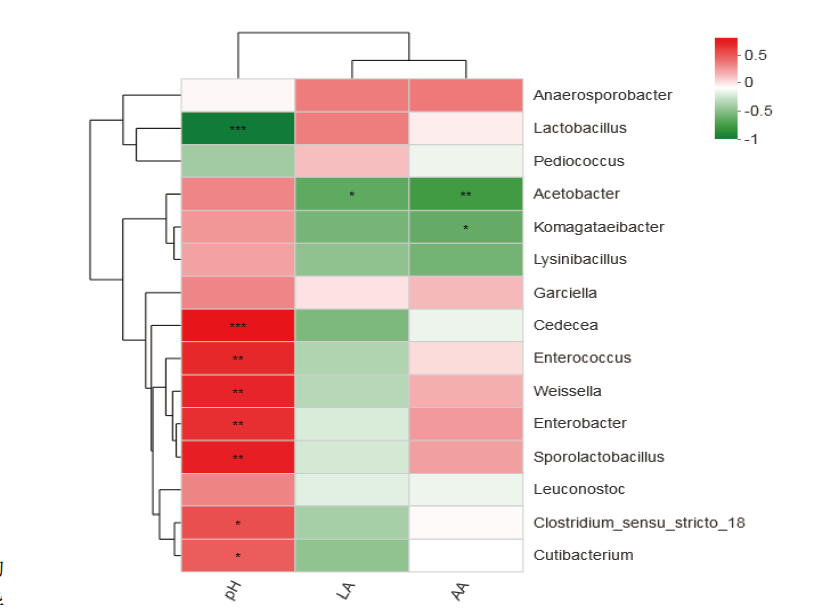
Fig. 8 Heatmap analysis of correlation between silage quality and bacterial community The X-axis and Y-axis are environmental factors and species,respectively,and the correlation R value and P value are obtained through calculation. The legend on the right is the color interval for different R values. * refers to 0.01<P≤0.05,** refers to 0.001<P≤0.01,*** refers to P≤0.001
| [1] |
Muck RE, Nadeau EMG, McAllister TA, et al. Silage review:Recent advances and future uses of silage additives[J]. J Dairy Sci, 2018, 101(5):3980-4000.
doi: S0022-0302(18)30322-9 pmid: 29685273 |
| [2] |
McAllister TA, Feniuk R, Mir Z, et al. Inoculants for alfalfa silage:Effects on aerobic stability, digestibility and the growth performance of feedlot steers[J]. Livest Prod Sci, 1998, 53(2):171-181.
doi: 10.1016/S0301-6226(97)00150-4 URL |
| [3] |
Dunière L, Sindou J, et al. Silage processing and strategies to prevent persistence of undesirable microorganisms[J]. Animal Feed Sci Technol, 2013, 182(1/2/3/4):1-15.
doi: 10.1016/j.anifeedsci.2013.04.006 URL |
| [4] |
Woolford MK. The detrimental effects of air on silage[J]. J Appl Bacteriol, 1990, 68(2):101-116.
doi: 10.1111/jam.1990.68.issue-2 URL |
| [5] | Basso FC, Lara EC, Assis F, et al. Fermentation characteristics and aerobic stability of corn silages inoculated with Bacillus subtilis[J]. Revista Brasileira de Saudee Producao Animal, 2012, 13(4):1009-1019. |
| [6] |
Hu ZF, Niu HX, Tong Q, et al. The microbiota dynamics of alfalfa silage during ensiling and after air exposure, and the metabolomics after air exposure are affected by Lactobacillus casei and cellulase addition[J]. Front Microbiol, 2020, 11:519121.
doi: 10.3389/fmicb.2020.519121 URL |
| [7] |
Filya I, Sucu E, Karabulut A. The effect of Lactobacillus buchneri on the fermentation, aerobic stability and ruminal degradability of maize silage[J]. J Appl Microbiol, 2006, 101(6):1216-1223.
pmid: 17105551 |
| [8] |
Mori H, Maruyama F, et al. Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes[J]. DNA Res, 2014, 21(2):217-227.
doi: 10.1093/dnares/dst052 URL |
| [9] |
Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur:open-source, platform-independent, community-supported software for describing and comparing microbial communities[J]. Appl Environ Microbiol, 2009, 75(23):7537-7541.
doi: 10.1128/AEM.01541-09 URL |
| [10] |
Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection[J]. Bioinformatics, 2011, 27(16):2194-2200.
doi: 10.1093/bioinformatics/btr381 URL |
| [11] |
Liu QH, Dong ZH, Shao T. Effect of additives on fatty acid profile of high moisture alfalfa silage during ensiling and after exposure to air[J]. Animal Feed Sci Technol, 2018, 236:29-38.
doi: 10.1016/j.anifeedsci.2017.11.022 URL |
| [12] |
Zhang L, Zhou X, Gu Q, et al. Analysis of the correlation between bacteria and fungi in sugarcane tops silage prior to and after aerobic exposure[J]. Bioresour Technol, 2019, 291:121835.
doi: 10.1016/j.biortech.2019.121835 URL |
| [13] |
Zhang T, Li L, Wang XF, et al. Effects of Lactobacillus buchneri and Lactobacillus plantarum on fermentation, aerobic stability, bacteria diversity and ruminal degradability of alfalfa silage[J]. World J Microbiol Biotechnol, 2009, 25(6):965-971.
doi: 10.1007/s11274-009-9973-x URL |
| [14] |
Li XM, Chen F, Wang XK, et al. Impacts of low temperature and ensiling period on the bacterial community of oat silage by SMRT[J]. Microorganisms, 2021, 9(2):274.
doi: 10.3390/microorganisms9020274 URL |
| [15] |
Li Y, Nishino N. Bacterial and fungal communities of wilted Italian ryegrass silage inoculated with and without Lactobacillus rhamnosus or Lactobacillus buchneri[J]. Lett Appl Microbiol, 2011, 52(4):314-321.
doi: 10.1111/j.1472-765X.2010.03000.x pmid: 21204884 |
| [16] | 何轶群, 雷赵民, 等. 青贮饲料中优良乳酸菌的分离鉴定及其生物学特性研究[J]. 生物技术通报, 2013(5):177-183. |
| He YQ, Lei ZM, et al. Isolation and identification of excellent lactic acid bacteria from silage and its biological characteristics research[J]. Biotechnol Bull, 2013(5):177-183. | |
| [17] |
Carvalho BF, Sales GFC, Schwan RF, et al. Criteria for lactic acid bacteria screening to enhance silage quality[J]. J Appl Microbiol, 2021, 130(2):341-355.
doi: 10.1111/jam.v130.2 URL |
| [18] |
Spoelstra SF, Courtin MG, Beers JACV. Acetic acid bacteria can initiate aerobic deterioration of whole crop maize silage[J]. J Agric Sci, 1988, 111(1):127-132.
doi: 10.1017/S0021859600082915 URL |
| [19] |
Jiang FG, Cheng HJ, Liu D, et al. Treatment of whole-plant corn silage with lactic acid bacteria and organic acid enhances quality by elevating acid content, reducing pH, and inhibiting undesirable microorganisms[J]. Front Microbiol, 2020, 11:593088.
doi: 10.3389/fmicb.2020.593088 URL |
| [1] | ZHONG Hui, LIU Ya-jun, WANG Bin-hua, HE Meng-jie, WU Lan. Effects of Analysis Methods on the Analyzed Results of 16S rRNA Gene Amplicon Sequencing in Bacterial Communities [J]. Biotechnology Bulletin, 2022, 38(6): 81-92. |
| [2] | XU Guang, WANG Meng-jiao, DENG Bai-wan, GUO Miao-miao. Bacterial Diversity and Community Structure of Rhizosphere Soil of Tea Plants in Different Years of Planting [J]. Biotechnology Bulletin, 2020, 36(3): 124-132. |
| [3] | WANG Yong-yan, ZHAO Bing-he, LIANG Guang-yu, LI Yun, XU Yang-cang. Bacterial Community Characteristics of Cultured Seawater with Microecological Preparations in Different Seasons [J]. Biotechnology Bulletin, 2020, 36(2): 126-133. |
| [4] | ZHANG Shi-wei, CHEN Xi, ZHONG Qi-ding, HUANG Zhan-bin, MENG Zhen, LUO Jin-xue, SHI Ling, BAI Zhi-hui. Microbial Communities on the Wine Grape Surfaces of Different Cultivars [J]. Biotechnology Bulletin, 2017, 33(3): 128-137. |
| [5] | WEI Guang-shan, ZHANG Jia-wei, LI Ming-cong, GAO Zheng. The Diversity and Distribution Pattern of Bacterial Community in the Water of Yellow River Estuary [J]. Biotechnology Bulletin, 2017, 33(10): 199-208. |
| [6] | Yu Rui,Zuo Fanglei, Chen Xiling, Wei Yanjie,Chen Shangwu. Introducing gshF into Lactobacillus paracasei L14 to Influence Its Stress Risistance Ability [J]. Biotechnology Bulletin, 2014, 0(9): 149-156. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||