








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (5): 112-122.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0946
Previous Articles Next Articles
XUE Xian-li1( ), WANG Jing-ran1, BI Hang-hang1, WANG De-pei1,2(
), WANG Jing-ran1, BI Hang-hang1, WANG De-pei1,2( )
)
Received:2021-07-24
Online:2022-05-26
Published:2022-06-10
Contact:
WANG De-pei
E-mail:xuexianli@tust.edu.cn;wangdp@tust.edu.cn
XUE Xian-li, WANG Jing-ran, BI Hang-hang, WANG De-pei. Effect of Spt7 Overexpression of on the Growth and Stress Resistance of Aspergillus niger[J]. Biotechnology Bulletin, 2022, 38(5): 112-122.
| 名称Name | 引物序列Sequence of primers(5'-3') | 引物作用Functions of primers |
|---|---|---|
| Pgla-F | TCACTACAGATACAAGGATCCTGCCATTGGCGGAGGGGT | pOEspt7质粒构建引物(pgla-spt7通过重组酶连接到p66质粒) |
| Pgla-R | AGCGACATTGCTGAGGTGTAATGATGCTGG | |
| spt7-F | TACACCTCAGCAATGTCGCTCGGACACCACC | |
| spt7-R(含终止子) | TGTGATCCGCCTGGAGGATCCCAAGGCAAGTACACTGAACAAGGA | |
| YZ-Ku70LL-F | CCGTCCGCCATAGTAGAATGT | pOE spt7质粒验证引物 |
| YZ-spt7-5R | GTTCTTACGAACCTCGCTCAT | |
| YZ-spt7-3R | AAACAACAACGAGCCTTTGGTC | |
| YZ-ku70RR-R | TTGTTGGATTCGCTAGTGCGCT | |
| RT-actin-F | ACCACCGACTCCCTACTA | pOE spt7 qRT-PCR引物 |
| RT-actin-R | AGTCAAGAGAGAGATGGGAT | |
| RT-hsp90-F | TCTTCCCTCTCTCTCTACCTC | |
| RT-hsp90-R | AAAACGGTTTGAAAGGTGTG | |
| RT-hsp70-F | GTGGGTGTCTTCCGTGATGACCGCA | |
| RT-hsp70-R | GGACCTCAGCGTCGGCAAA | |
| RT-hsp40-F | TGTTCAAGAAACCCAACATCCT | |
| RT-hsp40-R | TGTGGGAGACGACGGCCATG | |
| RT-GPX-F | CGCCTTCTCAATCTCCTTCA | |
| RT-GPX-R | AGCGCATCAAGTGGAACTTT | |
| RT-SOD-F | TTATCGCTCACAATGGCTGCTT | |
| RT-SOD-R | GACAGGTCAGGGAGAGTAGC | |
| RT-catR-F | CTTGTCACCGAGTGCCCGTTT | |
| RT-catR-R | GTAATCCGGACCCTCCTGTTGGG | |
| RT-cpeB-F | AACGGTTCGCTCCTCTTAAC | |
| RT-cpeB-R | AGGAAATCTTTGCTCCGTACTT |
Table 1 Primers used in the experiment
| 名称Name | 引物序列Sequence of primers(5'-3') | 引物作用Functions of primers |
|---|---|---|
| Pgla-F | TCACTACAGATACAAGGATCCTGCCATTGGCGGAGGGGT | pOEspt7质粒构建引物(pgla-spt7通过重组酶连接到p66质粒) |
| Pgla-R | AGCGACATTGCTGAGGTGTAATGATGCTGG | |
| spt7-F | TACACCTCAGCAATGTCGCTCGGACACCACC | |
| spt7-R(含终止子) | TGTGATCCGCCTGGAGGATCCCAAGGCAAGTACACTGAACAAGGA | |
| YZ-Ku70LL-F | CCGTCCGCCATAGTAGAATGT | pOE spt7质粒验证引物 |
| YZ-spt7-5R | GTTCTTACGAACCTCGCTCAT | |
| YZ-spt7-3R | AAACAACAACGAGCCTTTGGTC | |
| YZ-ku70RR-R | TTGTTGGATTCGCTAGTGCGCT | |
| RT-actin-F | ACCACCGACTCCCTACTA | pOE spt7 qRT-PCR引物 |
| RT-actin-R | AGTCAAGAGAGAGATGGGAT | |
| RT-hsp90-F | TCTTCCCTCTCTCTCTACCTC | |
| RT-hsp90-R | AAAACGGTTTGAAAGGTGTG | |
| RT-hsp70-F | GTGGGTGTCTTCCGTGATGACCGCA | |
| RT-hsp70-R | GGACCTCAGCGTCGGCAAA | |
| RT-hsp40-F | TGTTCAAGAAACCCAACATCCT | |
| RT-hsp40-R | TGTGGGAGACGACGGCCATG | |
| RT-GPX-F | CGCCTTCTCAATCTCCTTCA | |
| RT-GPX-R | AGCGCATCAAGTGGAACTTT | |
| RT-SOD-F | TTATCGCTCACAATGGCTGCTT | |
| RT-SOD-R | GACAGGTCAGGGAGAGTAGC | |
| RT-catR-F | CTTGTCACCGAGTGCCCGTTT | |
| RT-catR-R | GTAATCCGGACCCTCCTGTTGGG | |
| RT-cpeB-F | AACGGTTCGCTCCTCTTAAC | |
| RT-cpeB-R | AGGAAATCTTTGCTCCGTACTT |

Fig.1 Structural and amino acid multiple sequence alignment of Spt7 protein and phylogenetic tree analysis A: Structure of Spt7 protein in the yeast and Aspergillus niger. B: Amino acid multiple sequence alignment of Spt7 protein in the yeast and Aspergillus niger. C: Phylogenetic tree of the filamentous fungus Spt7
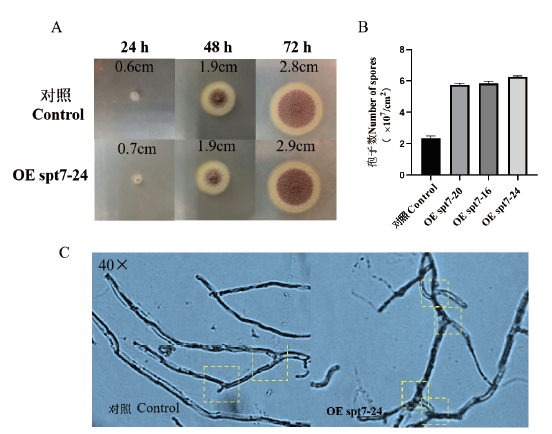
Fig. 2 Growth morphology observation and spore count of control and transformants at 30℃ A: Control group and OE spt7 colony morphology. B: Count of spores. C: Mycelial morphology of control and OE spt7 transformants under 40× microscope
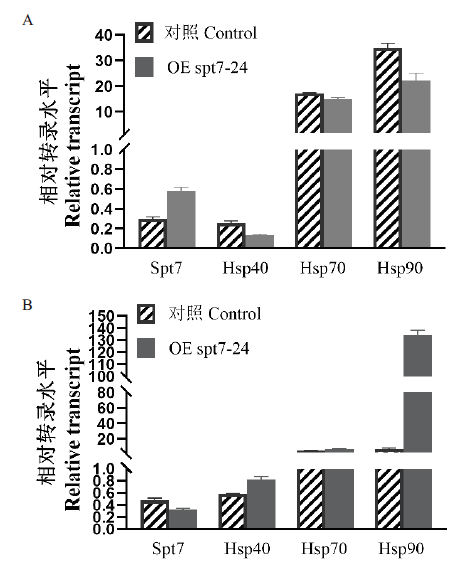
Fig. 7 Transcript levels of intracellular heat shock proteins in the control and OE spt7 transformants under heat stress conditions at 42℃ A:Transcript levels of heat shock protein after 3 h of heat stress. B:Transcript levels of heat shock protein after 5 h of heat stress
| [1] |
Soffers JHM, Workman JL. The SAGA chromatin-modifying complex:the sum of its parts is greater than the whole[J]. Genes Dev, 2020, 34(19/20):1287-1303.
doi: 10.1101/gad.341156.120 URL |
| [2] |
Hellman LM, Fried MG. Electrophoretic mobility shift assay(EMSA)for detecting protein-nucleic acid interactions[J]. Nat Protoc, 2007, 2(8):1849-1861.
pmid: 17703195 |
| [3] |
Prasad V, Voigt A. Quantification of protein aggregates using bimolecular fluorescence complementation[J]. Methods Mol Biol, 2019, 1873:183-193.
doi: 10.1007/978-1-4939-8820-4_11 pmid: 30341610 |
| [4] | Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast[J]. Nat Rev Mol Cell Biol, 2003, 4(4):276-284. |
| [5] |
Morgan MT, Haj-Yahya M, Ringel AE, et al. Structural basis for histone H2B deubiquitination by the SAGA DUB module[J]. Science, 2016, 351(6274):725-728.
doi: 10.1126/science.aac5681 URL |
| [6] |
Sung MK, Huh WK. In vivo quantification of protein-protein interactions in Saccharomyces cerevisiae using bimolecular fluorescence complementation assay[J]. J Microbiol Methods, 2010, 83(2):194-201.
doi: 10.1016/j.mimet.2010.08.021 URL |
| [7] |
Doncheva NT, Morris JH, Gorodkin J, et al. Cytoscape StringApp:network analysis and visualization of proteomics data[J]. J Proteome Res, 2019, 18(2):623-632.
doi: 10.1021/acs.jproteome.8b00702 URL |
| [8] |
VanHook AM. Linking light to development and metabolism[J]. Sci Signal, 2008, 1(24):ec222. DOI: 10.1126/scisignal.124ec222.
doi: 10.1126/scisignal.124ec222 |
| [9] | Moraga F, Aquea F. Composition of the SAGA complex in plants and its role in controlling gene expression in response to abiotic stresses[J]. Front Plant Sci, 2015, 6:865. |
| [10] |
Wu PY, Winston F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex[J]. Mol Cell Biol, 2002, 22(15):5367-5379.
doi: 10.1128/MCB.22.15.5367-5379.2002 URL |
| [11] |
Winston F, Dollard C, Malone EA, et al. Three genes are required for trans-activation of Ty transcription in yeast[J]. Genetics, 1987, 115(4):649-656.
doi: 10.1093/genetics/115.4.649 pmid: 3034719 |
| [12] |
Gansheroff LJ, Dollard C, Tan P, et al. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo[J]. Genetics, 1995, 139(2):523-536.
doi: 10.1093/genetics/139.2.523 pmid: 7713415 |
| [13] |
Papai G, Frechard A, Kolesnikova O, et al. Structure of SAGA and mechanism of TBP deposition on gene promoters[J]. Nature, 2020, 577(7792):711-716.
doi: 10.1038/s41586-020-1944-2 URL |
| [14] |
Wang H, Dienemann C, Stützer A, et al. Structure of the transcription coactivator SAGA[J]. Nature, 2020, 577(7792):717-720.
doi: 10.1038/s41586-020-1933-5 URL |
| [15] |
Li X, Seidel CW, Szerszen LT, et al. Enzymatic modules of the SAGA chromatin-modifying complex play distinct roles in Drosophila gene expression and development[J]. Genes Dev, 2017, 31(15):1588-1600.
doi: 10.1101/gad.300988.117 URL |
| [16] | Torres-Zelada EF, Weake VM. The Gcn5 complexes in Drosophila as a model for Metazoa[J]. Biochim et Biophys Acta BBA Gene Regul Mech, 2021, 1864(2):194610. |
| [17] |
Grasser KD, Rubio V, Barneche F. Multifaceted activities of the plant SAGA complex[J]. Biochim Biophys Acta Gene Regul Mech, 2021, 1864(2):194613.
doi: 10.1016/j.bbagrm.2020.194613 URL |
| [18] |
Mustachio LM, Roszik J, Farria A, et al. Targeting the SAGA and ATAC transcriptional coactivator complexes in MYC-driven cancers[J]. Cancer Res, 2020, 80(10):1905-1911.
doi: 10.1158/0008-5472.CAN-19-3652 pmid: 32094302 |
| [19] |
Wang L, Dent SY. Functions of SAGA in development and disease[J]. Epigenomics, 2014, 6(3):329-339.
doi: 10.2217/epi.14.22 pmid: 25111486 |
| [20] |
Nguyen KT, Ho QN, Pham TH, et al. The construction and use of versatile binary vectors carrying pyrG auxotrophic marker and fluorescent reporter genes for Agrobacterium-mediated transformation of Aspergillus oryzae[J]. World J Microbiol Biotechnol, 2016, 32(12):1-9.
doi: 10.1007/s11274-015-1971-6 URL |
| [21] | 曹张磊, 王德培, 张岚. 提高根癌农杆菌介导黑曲霉转化效率的研究[J]. 天津科技大学学报, 2016, 31(2):20-25. |
| Cao ZL, Wang DP, Zhang L. Improvement of transformation efficiency of Aspergillus niger mediated by Agrobacterium tumefaciens[J]. J Tianjin Univ Sci Technol, 2016, 31(2):20-25. | |
| [22] | 周磊, 韩一帆, 段志强. 含溴结构域蛋白2结构与功能的研究进展[J]. 中国细胞生物学学报, 2021, 43(4):856-865. |
| Zhou L, Han YF, Duan ZQ. Advances in the structure and function of bromodomain-containing protein 2[J]. Chin J Cell Biol, 2021, 43(4):856-865. | |
| [23] | 魏松红, 刘伟, 王海宁, 等. 中国植物病理学会2017年学术年会论文集[C]. 泰安: 中国植物病理学会, 2017. |
| Wei SH, Liu W, Wang HN, et al Proceedings of the 2017 Annual Academic Conference of the Chinese Society of Phytopathology[C]. Tai’an: Chinese Society of Plant Pathology, 2017. | |
| [24] | 包鹏飞, 周敏芬, 汤璧嘉, 等. 黏膜上皮细胞对高渗盐水的耐受性研究[J]. 嘉兴学院学报, 2010, 22(6):29-33. |
| Bao PF, Zhou MF, Tang BJ, et al. On tolerance of epithelial cells in hypertonic saline[J]. J Jiaxing Univ, 2010, 22(6):29-33. | |
| [25] |
Jenkins DE, Schultz JE, Matin A. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli[J]. J Bacteriol, 1988, 170(9):3910-3914.
pmid: 3045081 |
| [26] |
Jenkins DE, Auger EA, Matin A. Role of RpoH, a heat shock regulator protein, in Escherichia coli carbon starvation protein synthesis and survival[J]. J Bacteriol, 1991, 173(6):1992-1996.
pmid: 2002001 |
| [27] | Nystroem M, Nuortila-Jokinen J. Current Trends in Membrane Technology[J]. Kemia. Kemi, 1996, 23(8):. 663-672. |
| [28] |
Nyström T. To be or not to be:the ultimate decision of the growth-arrested bacterial cell[J]. FEMS Microbiol Rev, 1998, 21(4):283-290.
doi: 10.1016/S0168-6445(97)00060-0 URL |
| [29] |
Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster[J]. Science, 1994, 263(5150):1128-1130.
pmid: 8108730 |
| [30] |
Levine RL. Oxidative modification of glutamine synthetase. II. Characterization of the ascorbate model system[J]. J Biol Chem, 1983, 258(19):11828-11833.
pmid: 6137484 |
| [31] |
Kirkman HN, Gaetani GF. Mammalian catalase:a venerable enzyme with new mysteries[J]. Trends Biochem Sci, 2007, 32(1):44-50.
pmid: 17158050 |
| [32] |
Wang Y, Branicky R, Noë A, et al. Superoxide dismutases:Dual roles in controlling ROS damage and regulating ROS signaling[J]. J Cell Biol, 2018, 217(6):1915-1928.
doi: 10.1083/jcb.201708007 URL |
| [33] | Halliwell B, Gutteridge JMC. Redox chemistry:the essentials[M]// Free Radicals in Biology and Medicine. Oxford: Oxford University Press, 2015:30-76. |
| [34] |
Liu XZ, Huang BR. Heat stress injury in relation to membrane lipid peroxidation in creeping bentgrass[J]. Crop Sci, 2000, 40(2):503-510.
doi: 10.2135/cropsci2000.402503x URL |
| [35] |
Xu QZ, Huang BR. Morphological and physiological characteristics associated with heat tolerance in creeping bentgrass[J]. Crop Sci, 2001, 41(1):127-133.
doi: 10.2135/cropsci2001.411127x URL |
| [36] |
Zhang XZ, Ervin EH. Impact of seaweed extract-based cytokinins and Zeatin riboside on creeping bentgrass heat tolerance[J]. Crop Sci, 2008, 48(1):364-370.
doi: 10.2135/cropsci2007.05.0262 URL |
| [37] |
Ostankovitch M, Buchner J. The network of molecular chaperones:insights in the cellular proteostasis machinery[J]. J Mol Biol, 2015, 427(18):2899-2903.
doi: 10.1016/j.jmb.2015.08.010 pmid: 26363891 |
| [38] |
Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis[J]. Nature, 2011, 475(7356):324-332.
doi: 10.1038/nature10317 URL |
| [39] | Scharf KD, Berberich T, Ebersberger I, et al. The plant heat stress transcription factor(Hsf)family:structure, function and evolution[J]. Biochim Biophys Acta, 2012, 1819(2):104-119. |
| [40] |
Wang WX, Vinocur B, Shoseyov O, et al. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response[J]. Trends Plant Sci, 2004, 9(5):244-252.
doi: 10.1016/j.tplants.2004.03.006 URL |
| [41] | 蒲力群, 王逢会, 霍满鹏. 热休克蛋白的研究进展[J]. 延安大学学报:自然科学版, 2008, 27(1):72-75. |
| Pu LQ, Wang FH, Huo MP. The study progress of heat shock protein[J]. J Yanan Univ:Nat Sci Ed, 2008, 27(1):72-75. |
| [1] | GUO Yu-fei, YAN Rong-mei, ZHANG Xiao-ru, CAO Wei, LIU Hao. Metabolic Engineering Modification of Aspergillus niger for the Production of D-glucaric Acid [J]. Biotechnology Bulletin, 2022, 38(11): 227-237. |
| [2] | MENG Xiao-jian, YU Jian-dong, ZHENG Xiao-mei, ZHENG Ping, LI Zhi-min, SUN Ji-bin, YE Qin. Regulations of Small-molecules Metabolites on Hexokinase and Pyruvate Kinase in Aspergillus niger [J]. Biotechnology Bulletin, 2021, 37(12): 180-190. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||