








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (12): 180-190.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0274
Previous Articles Next Articles
MENG Xiao-jian1,2,3( ), YU Jian-dong2,3, ZHENG Xiao-mei2,3,4(
), YU Jian-dong2,3, ZHENG Xiao-mei2,3,4( ), ZHENG Ping2,3,4, LI Zhi-min1(
), ZHENG Ping2,3,4, LI Zhi-min1( ), SUN Ji-bin2,3,4, YE Qin1
), SUN Ji-bin2,3,4, YE Qin1
Received:2021-03-10
Online:2021-12-26
Published:2022-01-19
Contact:
ZHENG Xiao-mei,LI Zhi-min
E-mail:mengxj@tib.cas.cn;zheng_xm@tib.cas.cn;lizm@ecust.edu.cn
MENG Xiao-jian, YU Jian-dong, ZHENG Xiao-mei, ZHENG Ping, LI Zhi-min, SUN Ji-bin, YE Qin. Regulations of Small-molecules Metabolites on Hexokinase and Pyruvate Kinase in Aspergillus niger[J]. Biotechnology Bulletin, 2021, 37(12): 180-190.
| Primer name | Primer sequence(5'-3') | Source |
|---|---|---|
| HxkA-F | gcggcggtggctcctctagaATGGTTGG- AATCGGTCCTAAG | This study |
| HxkA-R | ccgtcgcggtcgactctagaTTATAGCA- GGGTCTTCATGTC | This study |
| PkiA-F | gcggcggtggctcctctagaATGGCCGC- CAGCTCTTCCC | This study |
| PkiA-R | ccgtcgcggtcgactctagaTTACTCAGC- CAGGCCAAG | This study |
| GV-F | CGGAGATTCGTCGCCTAATGTC | This study |
| GV-R | CCGTCGGTCGCAATACAATCAC | This study |
Table 1 Strains,plasmids and primers used in this study
| Primer name | Primer sequence(5'-3') | Source |
|---|---|---|
| HxkA-F | gcggcggtggctcctctagaATGGTTGG- AATCGGTCCTAAG | This study |
| HxkA-R | ccgtcgcggtcgactctagaTTATAGCA- GGGTCTTCATGTC | This study |
| PkiA-F | gcggcggtggctcctctagaATGGCCGC- CAGCTCTTCCC | This study |
| PkiA-R | ccgtcgcggtcgactctagaTTACTCAGC- CAGGCCAAG | This study |
| GV-F | CGGAGATTCGTCGCCTAATGTC | This study |
| GV-R | CCGTCGGTCGCAATACAATCAC | This study |
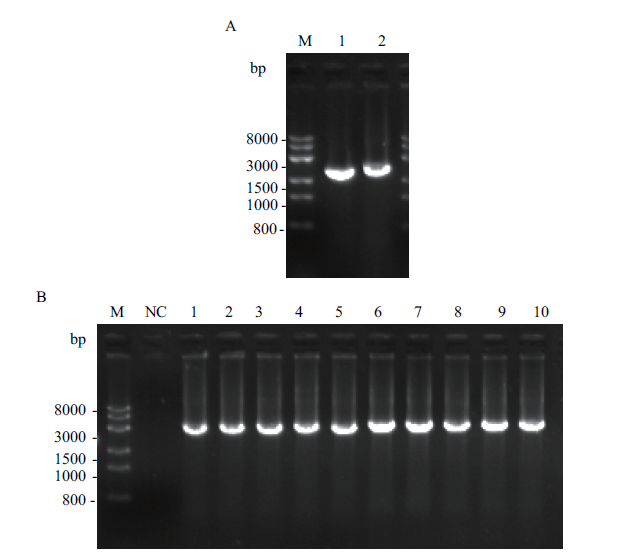
Fig. 1 Electrophoretogram of PCR products used for the construction of hexokinase(HxkA)and pyruvate kinase(PkiA)expressed A. niger strains A:PCR products of DNA sequences of hexokinase gene HxkA and pyruvate kinase gene PkiA. M:Trans 8K DNA marker. 1:HxkA gene. 2:PkiA gene. B:Genome PCR verification of recombinant expressing A. niger strain AnG1-HxkA and AnG1-PkiA. M:Trans 8K DNA marker. NC:Negative control. 1-5:The genome PCR verification of the transformants of AnG1-HxkA. 6-10:The genome PCR verification of the transformants of AnG1-PkiA

Fig.2 SDS-PAGE of the recombinant GST-fused HxkA and PkiA proteins before and after protein purification by affinity chromatography A:SDS-PAGE of GST-HxkA before and after affinity chromatography. M:Protein marker. 1:SDS-PAGE of GST-HxkA before affinity chromatography. 2:SDS-PAGE of recombinant GST-HxkA after affinity chromatography. B:SDS-PAGE of GST-PkiA before and after affinity chromatography. M:Protein marker. 1:SDS-PAGE of GST-PkiA before affinity chromatography. 2:SDS-PAGE of GST-PkiA after affinity chromatography
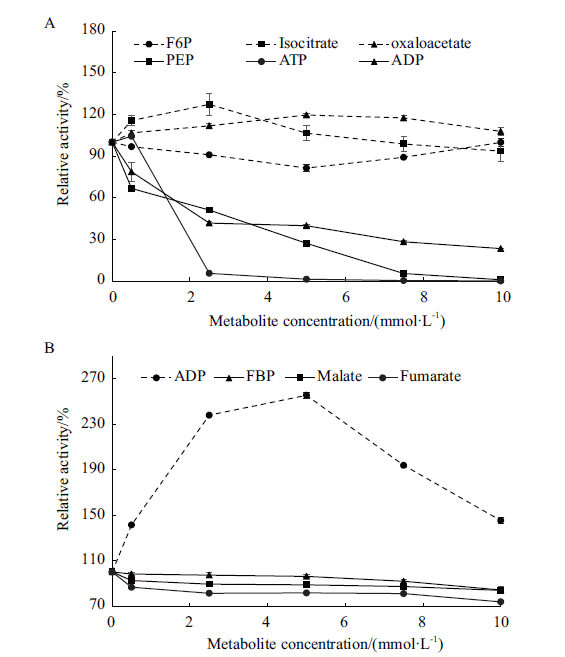
Fig.4 Effects of key metabolites under different concent-rations on the activities of HxkA(A)and PkiA(B)in A. niger The dotted lines refer to changes in enzyme activity in the presence of small molecule metabolites with activation at different concentrations. Solid lines refer to changes in enzyme activity in the presence of small molecule metabolites with inhibition at different concentrations

Fig.5 Multiple sequence alignment of HxkA and PkiA from A. niger A:Multiple sequence alignment of HxkA from A. niger with hexokinases from different species,the substrate binding sites of hexokinases are labeled with red stars.B:Multiple sequence alignment of PkiA from A. niger with pyruvate kinases from different species,the catalytic active sites of pyruvate kinases are labeled with red stars

Fig.6 Simulated 3D structure results of HxkA(A)and PkiA(B)from A. niger based on homologous modeling A:Protein structures of 2 sub-units of HxkA in A. niger shown in blue and green;the catalytic active sites of HxkA were labeled in red. B:Protein structures of 4 sub-units of PkiA shown in blue,green,purple and yellow;the catalytic active sites of PkiA are labeled in red

Fig.7 Molecular docking simulation of HxkA with key metabolites A-D:Molecular docking simulation of HxkA with fructose 6-phosphate,PEP,ADP,and ATP,respectively. The hydrogen bonds between the amino acids and key metabolites are represented as yellow dash lines,the hydrogen bond distances are represented in red numbers
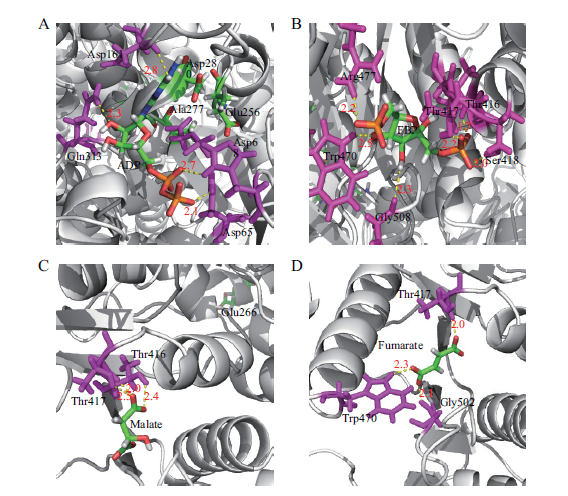
Fig.8 Molecular docking simulation of PkiA and key metabolites A-D:Molecular docking simulation of PkiA with ADP,fructose 1,6-diphosphate,malic acid,and fumaric acid,respectively. The hydrogen bonds between the amino acids and key metabolites are represented as yellow dash lines;the hydrogen bond distances are represented in red numbers.
| [1] |
Tong Z, Zheng X, Tong Y, et al. Systems metabolic engineering for citric acid production by Aspergillus niger in the post-genomic era[J]. Microb Cell Fact, 2019, 18(1):28.
doi: 10.1186/s12934-019-1064-6 URL |
| [2] |
Anastassiadis S, Morgunov IG, Kamzolova SV, et al. Citric acid production patent review[J]. Recent Patents Biotechnol, 2008, 2(2):107-123.
doi: 10.2174/187220808784619757 URL |
| [3] |
Karaffa L, Kubicek CP. Aspergillus niger citric acid accumulation:do we understand this well working black box?[J]. Appl Microbiol Biotechnol, 2003, 61(3):189-196.
doi: 10.1007/s00253-002-1201-7 URL |
| [4] |
Legisa M, Mattey M. Changes in primary metabolism leading to citric acid overflow in Aspergillus niger[J]. Biotechnol Lett, 2007, 29(2):181-190.
pmid: 17120089 |
| [5] |
Papagianni M. Advances in citric acid fermentation by Aspergillus niger:biochemical aspects, membrane transport and modeling[J]. Biotechnol Adv, 2007, 25(3):244-263.
pmid: 17337335 |
| [6] |
Kubicek-Pranz EM, Mozelt M, Rohr M, et al. Changes in the concentration of fructose 2, 6-bisphosphate in Aspergillus niger during stimulation of acidogenesis by elevated sucrose concentration[J]. Biochim Biophys Acta, 1990, 1033(3):250-255.
pmid: 2156568 |
| [7] |
Mesojednik S, Legisa M. Posttranslational modification of 6-phosphofructo-1-kinase in Aspergillus niger[J]. Appl Environ Microbiol, 2005, 71(3):1425-1432.
doi: 10.1128/AEM.71.3.1425-1432.2005 URL |
| [8] |
Panneman H, Ruijter GJ, van den Broeck HC, et al. Cloning and biochemical characterisation of Aspergillus niger hexokinase——the enzyme is strongly inhibited by physiological concentrations of trehalose 6-phosphate[J]. Eur J Biochem, 1998, 258(1):223-232.
pmid: 9851713 |
| [9] |
Arisan-Atac I, Wolschek MF, Kubicek CP. Trehalose-6-phosphate synthase A affects citrate accumulation by Aspergillus niger under conditions of high glycolytic flux[J]. FEMS Microbiol Lett, 1996, 140(1):77-83.
pmid: 8666204 |
| [10] |
Meixner-Monori B, Kubicek CP, Röhr M. Pyruvate kinase from Aspergillus niger:a regulatory enzyme in glycolysis?[J]. Can J Microbiol, 1984, 30(1):16-22.
pmid: 6713301 |
| [11] |
Zheng X, Zheng P, Zhang K, et al. 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger[J]. ACS Synth Biol, 2019, 8(7):1568-1574.
doi: 10.1021/acssynbio.7b00456 URL |
| [12] | Ruijter GJG, Panneman H, Visser J. Overexpression of phosphofructokinase and pyruvate kinase in citric acid-producing Aspergillus niger[J]. Biochim et Biophys Acta, 1997, 1334(2/3):317-326. |
| [13] |
Kuettner EB, Kettner K, Keim A, et al. Crystal structure of hexokinase KlHxk1 of Kluyveromyces lactis:a molecular basis for understanding the control of yeast hexokinase functions via covalent modification and oligomerization[J]. J Biol Chem, 2010, 285(52):41019-41033.
doi: 10.1074/jbc.M110.185850 URL |
| [14] |
Jurica MS, Mesecar A, Heath PJ, et al. The allosteric regulation of pyruvate kinase by fructose-1, 6-bisphosphate[J]. Structure, 1998, 6(2):195-210.
pmid: 9519410 |
| [15] | Land H, Humble MS. YASARA:a tool to obtain structural guidance in biocatalytic investigations[M]. Protein Eng, 2018, 1685:43-67. |
| [16] |
Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server[J]. Nucleic Acids Res, 2014, 42(web server issue):W320-W324.
doi: 10.1093/nar/gku316 URL |
| [17] |
Xu Y, Zhou Y, Cao W, et al. Improved production of malic acid in Aspergillus niger by abolishing citric acid accumulation and enhancing glycolytic flux[J]. ACS Synth Biol, 2020, 9(6):1418-1425.
doi: 10.1021/acssynbio.0c00096 URL |
| [18] |
Mlakar T, Legisa M. Citrate inhibition-resistant form of 6-phosphofructo-1-kinase from Aspergillus niger[J]. Appl Environ Microbiol, 2006, 72(7):4515-4521.
doi: 10.1128/AEM.00539-06 URL |
| [19] |
Yuan M, McNae IW, Chen Y, et al. An allostatic mechanism for M2 pyruvate kinase as an amino-acid sensor[J]. Biochem J, 2018, 475(10):1821-1837.
doi: 10.1042/BCJ20180171 URL |
| [20] |
Susan-Resiga D, Nowak T. Proton donor in yeast pyruvate kinase:chemical and kinetic properties of the active site Thr 298 to Cys mutant[J]. Biochemistry, 2004, 43(48):15230-15245.
pmid: 15568816 |
| [21] |
Bond CJ, Jurica MS, Mesecar A, et al. Determinants of allosteric activation of yeast pyruvate kinase and identification of novel effectors using computational screening[J]. Biochemistry, 2000, 39(50):15333-15343.
pmid: 11112519 |
| [1] | CHENG Ting, YUAN Shuai, ZHANG Xiao-yuan, LIN Liang-cai, LI Xin, ZHANG Cui-ying. Research Progress in the Regulation of Isobutanol Synthesis Pathway in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(7): 80-90. |
| [2] | DUAN Yue-tong, WANG Peng-nian, ZHANG Chun-bao, LIN Chun-jing. Research Progress in Plant Flavanone-3-hydroxylase Gene [J]. Biotechnology Bulletin, 2022, 38(6): 27-33. |
| [3] | XUE Xian-li, WANG Jing-ran, BI Hang-hang, WANG De-pei. Effect of Spt7 Overexpression of on the Growth and Stress Resistance of Aspergillus niger [J]. Biotechnology Bulletin, 2022, 38(5): 112-122. |
| [4] | TIAN Qing-yin, YUE Yuan-zheng, SHEN Hui-min, PAN Duo, YANG Xiu-lian, WANG Liang-gui. Research Progress in the Regulation of Carotenoid Metabolism in Plant Ornamental Organs [J]. Biotechnology Bulletin, 2022, 38(12): 35-46. |
| [5] | GUO Yu-fei, YAN Rong-mei, ZHANG Xiao-ru, CAO Wei, LIU Hao. Metabolic Engineering Modification of Aspergillus niger for the Production of D-glucaric Acid [J]. Biotechnology Bulletin, 2022, 38(11): 227-237. |
| [6] | YUAN Kai, HE Wei, YANG Yun-li, ZHU Wei-yu, PENG Chao, AN Tai, LI Li, ZHOU Wei-qiang. Research Progress on Biosynthesis and Metabolic Regulation of Ganoderic Acids [J]. Biotechnology Bulletin, 2021, 37(8): 46-54. |
| [7] | MA qin, LEI Rui-feng, Dilireba Abudourousuli, Muyesaier Aosiman, Zulihumaer Rouzi, AN Deng-di. Research Progress on the Symbiotic Metabolic of Endophytes and Plants Under Stress [J]. Biotechnology Bulletin, 2021, 37(3): 153-161. |
| [8] | GAO Yue, GUO Xiao-peng, YANG Yang, ZHANG Miao-miao, LI Wen-jian, LU Dong. Research Progress of Biobutanol Fermentation [J]. Biotechnology Bulletin, 2018, 34(8): 27-34. |
| [9] | ZHENG Chen-hua, DU Xi-ping, LI Li-jun, LI Tian-li, CAO Ying, NI Hui. The Relationship Between Characteristics of Yielding Carotenoid and Expression of Carotenogenic Genes in Phaffia rhodozyma [J]. Biotechnology Bulletin, 2016, 32(2): 123-130. |
| [10] | CHEN Ke, DING Yan-ping, WANG Jian-lin, SHAO Bao-ping. Research Progress on p53-involved Metabolic Regulation [J]. Biotechnology Bulletin, 2016, 32(11): 52-58. |
| [11] | Zhang Xiaoli, Zheng Xiaomei, Man Yun, Luo Hu, Yu Jiandong, Zheng Ping, Liu Hao, Sun Jibin. Preparation of Protoplast for Efficient DNA Transformation of Citric Acid Hyper-producing Aspergillus niger Industrial Strain [J]. Biotechnology Bulletin, 2015, 31(3): 171-177. |
| [12] | Li Yawei Zhao Lin Hao Yongwei Tang Xiuli Yang Jikun Gao Yuzhen Liu Xinli . Screening and Identification of Interspecies Microorganisms Inducing Overproduction of Epothiones in Sorangium cellulosum [J]. Biotechnology Bulletin, 2013, 0(10): 137-141. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||