








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 204-215.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1066
Previous Articles Next Articles
YAN Hui-lin1( ), LU Guang-xin1(
), LU Guang-xin1( ), DENG Ye2, GU Song-song2, YAN Cheng-liang2, MA Kun1, ZHAO Yang-an1, ZHANG Hai-juan1, WANG Ying-cheng1, ZHOU Xue-li3, DOU Sheng-yun3
), DENG Ye2, GU Song-song2, YAN Cheng-liang2, MA Kun1, ZHAO Yang-an1, ZHANG Hai-juan1, WANG Ying-cheng1, ZHOU Xue-li3, DOU Sheng-yun3
Received:2021-08-19
Online:2022-10-26
Published:2022-11-11
Contact:
LU Guang-xin
E-mail:1016632996@qq.com;lugx74@qq.com
YAN Hui-lin, LU Guang-xin, DENG Ye, GU Song-song, YAN Cheng-liang, MA Kun, ZHAO Yang-an, ZHANG Hai-juan, WANG Ying-cheng, ZHOU Xue-li, DOU Sheng-yun. Effects of Rhizobium Seed Dressing on the Soil Microbial Community of Grass-legume Mixtures in Alpine Regions[J]. Biotechnology Bulletin, 2022, 38(10): 204-215.
| 牧草种类 Forage species | 混播草地 Mixed pasture | 重复数 Repetition | 标记 Remark |
|---|---|---|---|
| ‘川草2号’老芒麦(Elymus sibiricus L.cv.‘Chuancao No.2’) | 苜蓿‘北林201’(拌种) Medicago sativa cv.‘Beilin201’(Rhizobium) | 3 | A1 |
| 苜蓿‘北林201’Medicago sativa cv.‘Beilin201’ | 3 | D1 | |
| ‘阿坝’垂穗披碱草(Elynus nutans Griseb.cv.‘Aba’) | 苜蓿‘北林201’(拌种)Medicago sativa cv.‘Beilin201’(Rhizobium) | 3 | A2 |
| 苜蓿‘北林201’Medicago sativa cv.‘Beilin201’ | 3 | D2 |
Table 1 Experimental design
| 牧草种类 Forage species | 混播草地 Mixed pasture | 重复数 Repetition | 标记 Remark |
|---|---|---|---|
| ‘川草2号’老芒麦(Elymus sibiricus L.cv.‘Chuancao No.2’) | 苜蓿‘北林201’(拌种) Medicago sativa cv.‘Beilin201’(Rhizobium) | 3 | A1 |
| 苜蓿‘北林201’Medicago sativa cv.‘Beilin201’ | 3 | D1 | |
| ‘阿坝’垂穗披碱草(Elynus nutans Griseb.cv.‘Aba’) | 苜蓿‘北林201’(拌种)Medicago sativa cv.‘Beilin201’(Rhizobium) | 3 | A2 |
| 苜蓿‘北林201’Medicago sativa cv.‘Beilin201’ | 3 | D2 |
| 环境因子Environmental factors | 变量Variables | 分组Group | ||||
|---|---|---|---|---|---|---|
| DB | DN | HB | HN | |||
| 理化性质 Soil physical-chemical property | 总氮TN/% | 1 805.37±293.06 | 1 814.36±152.19 | 1 676.88±248.44 | 1 777.86±137.70 | |
| 铵态氮NH4-N /(mg·kg-1) | 4.41±1.11 | 4.22±0.54 | 4.52±0.94 | 4.30±1.00 | ||
| 硝态氮NO3-N /(mg·kg-1) | 38.87±2.85 | 39.97±1.70 | 41.84±4.35 | 40.06±2.08 | ||
| 有机碳TOC/% | 2.29±0.30 | 2.24±0.29 | 2.23±0.31 | 2.17±0.28 | ||
| 土壤性质 Soil property | 温度MAT/℃ | 13.49±3.54 | 11.88±0.18 | 13.98±4.36 | 11.87±0.20 | |
| 水分SMC/% | 9.81±1.06 | 10.63±1.31 | 10.63±1.23 | 10.70±1.45 | ||
| 电导率EC/(us·cm-1) | 0.06±0.04 | 0.04±0.02 | 0.07±0.0.1 | 0.04±0.02 | ||
Table 2 Soil physical and chemical properties
| 环境因子Environmental factors | 变量Variables | 分组Group | ||||
|---|---|---|---|---|---|---|
| DB | DN | HB | HN | |||
| 理化性质 Soil physical-chemical property | 总氮TN/% | 1 805.37±293.06 | 1 814.36±152.19 | 1 676.88±248.44 | 1 777.86±137.70 | |
| 铵态氮NH4-N /(mg·kg-1) | 4.41±1.11 | 4.22±0.54 | 4.52±0.94 | 4.30±1.00 | ||
| 硝态氮NO3-N /(mg·kg-1) | 38.87±2.85 | 39.97±1.70 | 41.84±4.35 | 40.06±2.08 | ||
| 有机碳TOC/% | 2.29±0.30 | 2.24±0.29 | 2.23±0.31 | 2.17±0.28 | ||
| 土壤性质 Soil property | 温度MAT/℃ | 13.49±3.54 | 11.88±0.18 | 13.98±4.36 | 11.87±0.20 | |
| 水分SMC/% | 9.81±1.06 | 10.63±1.31 | 10.63±1.23 | 10.70±1.45 | ||
| 电导率EC/(us·cm-1) | 0.06±0.04 | 0.04±0.02 | 0.07±0.0.1 | 0.04±0.02 | ||
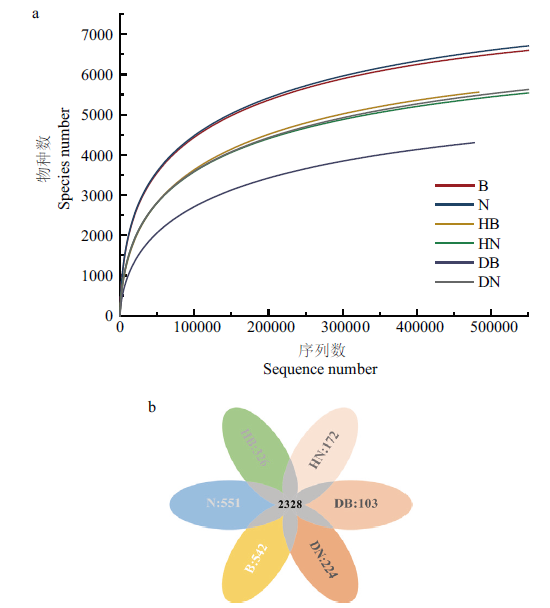
Fig. 2 Rarefaction curves for soil prokaryotic microbial communities and Venn diagram showing the unique and shared OTUs a: Sequencing depth has basically satisfied downstream analysis. b: Visualization of common and unique OTUs in different groups. B: Bulk soil seed dressing. N: Bulk soil without seed dressing. The same below
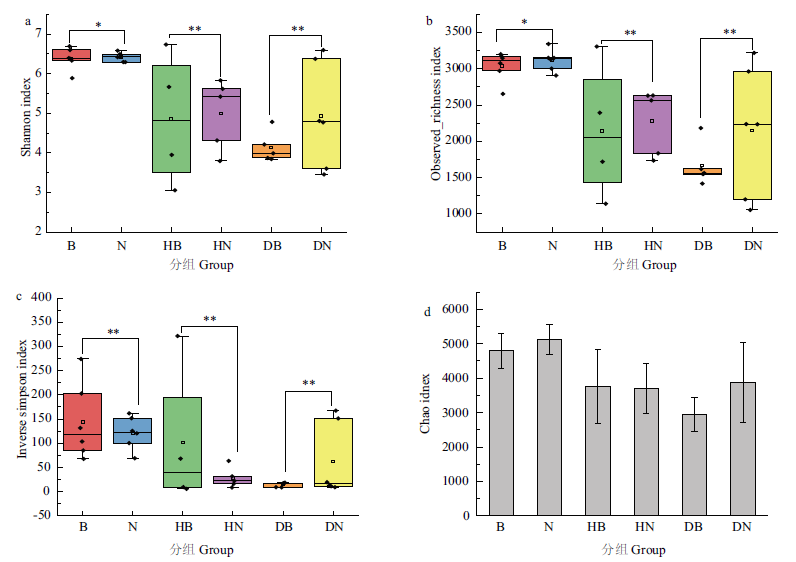
Fig. 3 α diversity analysis a:Shannon index analysis. b:Observed_richness index analysis. c:Inverse Simpson index analysis. d:Chao index analysis. * indicates significant differences,* P<0.05,** P<0.01,and *** P<0.001. The same below
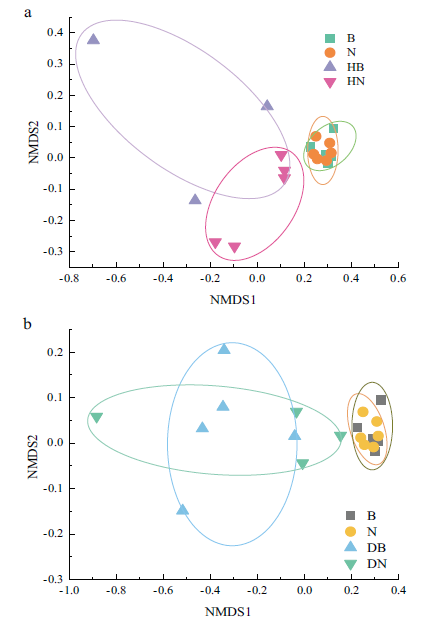
Fig. 5 NMDS of bulk and rhizosphere soil prokaryotic community structure a:Gramineous NMDS based on Bary-Curtis distance. b:Legume NMDS based on Bary-Curtis distance
| 整体比较Global test | 两两比较Pairwise composition | |||||||
|---|---|---|---|---|---|---|---|---|
| B | N | HB | HN | DB | DN | |||
| MRPP | 0.415 8 | B | 0.6 | 0.048 | 0.011 | 0.006 | 0.025 | |
| δ(P-value) | 0.001 | N | 0.291 6 | 0.004 | 0.002 | 0.004 | 0.008 | |
| HB | 0.462 1 | 0.461 0 | 0.75 | 0.504 | 0.946 | |||
| HN | 0.359 8 | 0.317 9 | 0.513 3 | 0.049 | 0.232 | |||
| DB | 0.362 1 | 0.320 3 | 0.516 1 | 0.392 8 | 0.368 | |||
| DN | 0.430 4 | 0.392 0 | 0.582 7 | 0.469 4 | 0.471 7 | |||
| ANOSIM | 0.423 6 | B | 0.73 | 0.011 | 0.012 | 0.003 | 0.03 | |
| R(P-value) | 0.001 | N | -0.055 | 0.008 | 0.002 | 0.001 | 0.009 | |
| HB | 0.488 0 | 0.678 5 | 0.284 | 0.185 | 0.606 | |||
| HN | 0.586 6 | 0.853 3 | 0.068 7 | 0.048 | 0.373 | |||
| DB | 0.936 | 0.997 3 | 0.131 2 | 0.384 | 0.48 | |||
| DN | 0.340 7 | 0.485 1 | -0.06 7 | 0.018 6 | -0.026 | |||
| PERMANOVA | 4.100 5 | B | 0.504 | 0.049 | 0.009 | 0.011 | 0.023 | |
| F(P-value) | 0.001 | N | 0.962 6 | 0.016 | 0.001 | 0.002 | 0.008 | |
| HB | 3.041 3 | 4.806 7 | 0.861 | 0.424 | 0.909 | |||
| HN | 6.581 5 | 11.153 1 | 0.541 9 | 0.045 | 0.286 | |||
| DB | 12.739 4 | 19.585 4 | 1.021 9 | 2.477 6 | 0.316 | |||
| DN | 4.704 1 | 7.176 4 | 0.416 5 | 1.189 4 | 1.150 2 | |||
Table 3 Dissimilarity test based on Bary-Curtis distance of soil prokaryotic communities
| 整体比较Global test | 两两比较Pairwise composition | |||||||
|---|---|---|---|---|---|---|---|---|
| B | N | HB | HN | DB | DN | |||
| MRPP | 0.415 8 | B | 0.6 | 0.048 | 0.011 | 0.006 | 0.025 | |
| δ(P-value) | 0.001 | N | 0.291 6 | 0.004 | 0.002 | 0.004 | 0.008 | |
| HB | 0.462 1 | 0.461 0 | 0.75 | 0.504 | 0.946 | |||
| HN | 0.359 8 | 0.317 9 | 0.513 3 | 0.049 | 0.232 | |||
| DB | 0.362 1 | 0.320 3 | 0.516 1 | 0.392 8 | 0.368 | |||
| DN | 0.430 4 | 0.392 0 | 0.582 7 | 0.469 4 | 0.471 7 | |||
| ANOSIM | 0.423 6 | B | 0.73 | 0.011 | 0.012 | 0.003 | 0.03 | |
| R(P-value) | 0.001 | N | -0.055 | 0.008 | 0.002 | 0.001 | 0.009 | |
| HB | 0.488 0 | 0.678 5 | 0.284 | 0.185 | 0.606 | |||
| HN | 0.586 6 | 0.853 3 | 0.068 7 | 0.048 | 0.373 | |||
| DB | 0.936 | 0.997 3 | 0.131 2 | 0.384 | 0.48 | |||
| DN | 0.340 7 | 0.485 1 | -0.06 7 | 0.018 6 | -0.026 | |||
| PERMANOVA | 4.100 5 | B | 0.504 | 0.049 | 0.009 | 0.011 | 0.023 | |
| F(P-value) | 0.001 | N | 0.962 6 | 0.016 | 0.001 | 0.002 | 0.008 | |
| HB | 3.041 3 | 4.806 7 | 0.861 | 0.424 | 0.909 | |||
| HN | 6.581 5 | 11.153 1 | 0.541 9 | 0.045 | 0.286 | |||
| DB | 12.739 4 | 19.585 4 | 1.021 9 | 2.477 6 | 0.316 | |||
| DN | 4.704 1 | 7.176 4 | 0.416 5 | 1.189 4 | 1.150 2 | |||
| 高水平表型 High-level phenotypes | 根周土 Bulk soil | 根际土 Rhizosphere soil | |||||
|---|---|---|---|---|---|---|---|
| B | N | HB | HN | DB | DN | ||
| 需氧Aerobic | 99 | 117 | 51 | 19 | 13 | 36 | |
| 厌氧Anaerobic | 48 | 50 | 30 | 24 | 13 | 30 | |
| 兼性厌氧 Facultatively_anaerobic | 25 | 18 | 14 | 9 | 3 | 10 | |
| 革兰氏阳性Gram positive | 39 | 48 | 116 | 84 | 39 | 102 | |
| 革兰氏阴性Gram_negative | 233 | 22 | 21 | 15 | 6 | 14 | |
Table 4 Bugbase results
| 高水平表型 High-level phenotypes | 根周土 Bulk soil | 根际土 Rhizosphere soil | |||||
|---|---|---|---|---|---|---|---|
| B | N | HB | HN | DB | DN | ||
| 需氧Aerobic | 99 | 117 | 51 | 19 | 13 | 36 | |
| 厌氧Anaerobic | 48 | 50 | 30 | 24 | 13 | 30 | |
| 兼性厌氧 Facultatively_anaerobic | 25 | 18 | 14 | 9 | 3 | 10 | |
| 革兰氏阳性Gram positive | 39 | 48 | 116 | 84 | 39 | 102 | |
| 革兰氏阴性Gram_negative | 233 | 22 | 21 | 15 | 6 | 14 | |
| [1] | 李成阳, 薛娴, 赖炽敏, 等. 青藏高原退化高寒草甸生长季承载力[J]. 中国沙漠, 2018, 38(6):1330-1338. |
| Li CY, Xue X, Lai CM, et al. Growing season bearing capacity of degraded alpine meadow in the Qinghai-Tibet plateau[J]. J Desert Res, 2018, 38(6):1330-1338. | |
| [2] | 苟文龙, 李平, 肖冰雪, 等. 禾豆牧草混播增产增效研究进展[J]. 草学, 2020(3):16-23. |
| Gou WL, Li P, Xiao BX, et al. Research status on grass-legume mixture[J]. Prataculture Animal Husb, 2020(3):16-23. | |
| [3] | 徐然然, 常生华, 贾倩民, 等. 施氮和利用方式对黄土高原禾豆混播草地产量、品质和水分利用的影响[J]. 草地学报, 2020, 28(6):1744-1755. |
| Xu RR, Chang SH, Jia QM, et al. Effects of nitrogen application and utilization methods on yield, quality and water use of grass-legume mixed grassland in loess plateau[J]. Acta Agrestia Sin, 2020, 28(6):1744-1755. | |
| [4] | 管凤贞, 邱宏端, 陈济琛, 等. 根瘤菌菌剂的研究与开发现状[J]. 生态学杂志, 2012, 31(3):755-759. |
| Guan FZ, Qiu HD, Chen JC, et al. Rhizobium inoculants:research progress and development status[J]. Chin J Ecol, 2012, 31(3):755-759. | |
| [5] | 常玮, 王炜, 屈新兰. 苜蓿根瘤菌菌剂的研究[J]. 新疆农业科学, 2004, 41(2):102-104. |
| Chang W, Wang W, Qu XL. Study on biologicals of Rhizobium meliloti dangeard[J]. Xinjiang Agric Sci, 2004, 41(2):102-104. | |
| [6] | 李桂花. 不同施肥对土壤微生物活性、群落结构和生物量的影响[J]. 中国农学通报, 2010, 26(14):204-208. |
| Li GH. Effect of organic amendments and chemical fertilizer on soil microbial activity, biomass and community structure[J]. Chin Agric Sci Bull, 2010, 26(14):204-208. | |
| [7] | 张燕. 不同土壤处理对设施土壤中微生物生物量的影响[J]. 中国农学通报, 2018, 34(26):90-98. |
| Zhang Y. Effects of different soil treatments on microbial biomass in facility soil[J]. Chin Agric Sci Bull, 2018, 34(26):90-98. | |
| [8] | 孟庆英, 张春峰, 于忠和, 等. 根瘤菌对大豆根际土壤微生物及大豆农艺性状的影响[J]. 大豆科学, 2012, 31(3):498-500. |
| Meng QY, Zhang CF, Yu ZH, et al. Effects of rhizobia on rhizosphere soil microoganisms and agronomic characters of soybean[J]. Soybean Sci, 2012, 31(3):498-500. | |
| [9] |
王孝林, 王二涛. 根际微生物促进水稻氮利用的机制[J]. 植物学报, 2019, 54(3):285-287.
doi: 10.11983/CBB19060 |
| Wang XL, Wang ET. NRT1. 1B connects root microbiota and nitrogen use in rice[J]. Chin Bull Bot, 2019, 54(3):285-287. | |
| [10] | Sharma R, Bisaria VS, Sharma S. Impact of rhizobial inoculants on rhizospheric microbial communities[M]// Hansen A, Choudhary D, Agrawal P, et al. Rhizobium Biology and Biotechnology, Springer, Cham, 2017:1-10. |
| [11] |
Cheng HY, Wu BD, Wei M, et al. Changes in community structure and metabolic function of soil bacteria depending on the type restoration processing in the degraded alpine grassland ecosystems in Northern Tibet[J]. Sci Total Environ, 2021, 755(Pt 2):142619.
doi: 10.1016/j.scitotenv.2020.142619 URL |
| [12] | Li JJ, Yang C, Zhou HK, et al. Responses of plant diversity and soil microorganism diversity to water and nitrogen additions in the Qinghai-Tibetan Plateau[J]. Glob Ecol Conserv, 2020, 22:e01003. |
| [13] |
Ren CJ, Zhang W, Zhong ZK, et al. Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics[J]. Sci Total Environ, 2018, 610/611:750-758.
doi: 10.1016/j.scitotenv.2017.08.110 URL |
| [14] |
Feng K, Zhang ZJ, Cai WW, et al. Biodiversity and species competition regulate the resilience of microbial biofilm community[J]. Mol Ecol, 2017, 26(21):6170-6182.
doi: 10.1111/mec.14356 pmid: 28926148 |
| [15] |
Magoč T, Salzberg SL. FLASH:fast length adjustment of short reads to improve genome assemblies[J]. Bioinformatics, 2011, 27(21):2957-2963.
doi: 10.1093/bioinformatics/btr507 URL |
| [16] |
Kong Y. Btrim:a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies[J]. Genomics, 2011, 98(2):152-153.
doi: 10.1016/j.ygeno.2011.05.009 URL |
| [17] |
DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB[J]. Appl Environ Microbiol, 2006, 72(7):5069-5072.
doi: 10.1128/AEM.03006-05 URL |
| [18] |
Edgar RC. UPARSE:highly accurate OTU sequences from microbial amplicon reads[J]. Nat Methods, 2013, 10(10):996-998.
doi: 10.1038/nmeth.2604 URL |
| [19] |
Tortosa G, AJ Fernández-González, Lasa A V, et al. Involvement of the metabolically active bacteria in the organic matter degradation during olive mill waste composting[J]. Science of The Total Environment, 2021, 789(40):147975.
doi: 10.1016/j.scitotenv.2021.147975 URL |
| [20] |
Ward T, Larson J, Meulemans J, et al. BugBase predicts organism-level microbiome phenotypes[J]. bioRxiv, 2017, DOI:10.1101/133462.
doi: 10.1101/133462 |
| [21] | 陈香碧, 苏以荣, 何寻阳, 等. 不同干扰方式对喀斯特生态系统土壤细菌优势类群—变形菌群落的影响[J]. 土壤学报, 2012, 49(2):354-363. |
| Chen XB, Su YR, He XY, et al. Effect of human disturbance on composition of the dominant bacterial group proteobacteriain Karst soil ecosystems[J]. Acta Pedol Sin, 2012, 49(2):354-363. | |
| [22] | 杜滢鑫, 谢宝明, 蔡洪生, 等. 大庆盐碱地九种植物根际土壤微生物群落结构及功能多样性[J]. 生态学报, 2016, 36(3):740-747. |
| Du YX, Xie BM, Cai HS, et al. Structural and functional diversity of rhizosphere microbial community of nine plant species in the Daqing Saline-alkali soil region[J]. Acta Ecol Sin, 2016, 36(3):740-747. | |
| [23] | 蔡秋华, 黄俊杰, 林云红, 等. 草木灰对烤烟根际土壤微生物群落结构及功能多样性的影响[J]. 中国农学通报, 2019, 35(11):43-50. |
| Cai QH, Huang JJ, Lin YH, et al. Plant ash:effect on rhizosphere microorganism population structure and functional diversity of flue-cured tobacco[J]. Chin Agric Sci Bull, 2019, 35(11):43-50. | |
| [24] | 刘长征, 周良云, 廖沛然, 等. 何首乌-穿心莲间作对何首乌根际土壤放线菌群落结构和多样性的影响[J]. 中国中药杂志, 2020, 45(22):5452-5458. |
| Liu CZ, Zhou LY, Liao PR, et al. Effect of Polygonum multiflorum-Andrographis paniculata intercropping system on rhizosphere soil actinomycetes community structure and diversity of P. multiflorum[J]. China J Chin Mater Med, 2020, 45(22):5452-5458. | |
| [25] |
Chakraborty U, Purkayastha RP. Role of rhizobitoxine in protecting soybean roots from Macrophomina phaseolina infection[J]. Can J Microbiol, 1984, 30(3):285-289.
pmid: 6539157 |
| [26] | 刘重喜. 抗部分植物病原真菌放线菌的分离及新种鉴定[D]. 哈尔滨: 东北农业大学, 2010. |
| Liu CX. Isolation of against some phytopathogenic fungi Actinomyces and identification of a novel actinomyce[D]. Harbin:Northeast Agricultural University, 2010. | |
| [27] | 喻岚晖, 王杰, 廖李容, 等. 青藏高原退化草甸土壤微生物量、酶化学计量学特征及其影响因素[J]. 草地学报, 2020, 28(6):1702-1710. |
| Yu LH, Wang J, Liao LR, et al. Soil microbial biomass, enzyme activities and ecological stoichiometric characteristics and influencing factors along degraded meadows on the Qinghai-Tibet plateau[J]. Acta Agrestia Sin, 2020, 28(6):1702-1710. | |
| [28] | 侯高礼. 土壤团粒结构及其促进形成[J]. 西北园艺:果树, 2013(1):52-53. |
| Hou GL. Soil aggregate structure and its promoting formation[J]. Northwest Hortic, 2013(1):52-53. | |
| [29] | 周文杰, 吕德国, 秦嗣军. 植物与根际微生物相互作用关系研究进展[J]. 吉林农业大学学报, 2016, 38(3):253-260. |
| Zhou WJ, Lv DG, Qin SJ. Research progress in interaction between plant and rhizosphere microorganism[J]. J Jilin Agric Univ, 2016, 38(3):253-260. | |
| [30] | 徐白璐. 长期施肥和温度对酸性旱地土壤硝化微生物和总细菌群落的影响[D]. 南京: 南京师范大学, 2017. |
| Xu BL. Effects of long-term fertilization and temperature on nitrifying microorganisms and total bacterial communities in acidic dryland soil[D]. Nanjing: Nanjing Normal University, 2017. | |
| [31] | Bano ANA. Effect of plant growth promoting rhizobacteria on root morphology of Safflower(Carthamus tinctorius L.)[J]. Afr J Biotechnol, 2011, 10(59):12639-12649. |
| [1] | CHEN Chu-wen, LI Jie, ZHAO Rui-peng, LIU Yuan, WU Jin-bo, LI Zhi-xiong. Cloning, Tissue Expression Profile and Function Prediction of GPX3 Gene in Tibetan Chicken [J]. Biotechnology Bulletin, 2023, 39(3): 311-320. |
| [2] | XIN Ya-fen, CHEN Chen, ZENG Tai-ru, DU Zhao-chang, NI Hao-ran, ZHONG Yi-hao, TAN Xiao-ping, YAN Yan-hong. Research Progress in the Effects of Additives to Silage on Microbial Diversity [J]. Biotechnology Bulletin, 2021, 37(9): 24-30. |
| [3] | WANG Ting, YANG Yang, LI Jin-ping, DU Kun. Research Progress in the Effects of Genetically Modified Crops on Soil Microbial Community [J]. Biotechnology Bulletin, 2021, 37(9): 255-265. |
| [4] | ZHANG Ying-chao, YIN Shou-liang, WANG Yi-wei, WANG Xue-kai, YANG Fu-yu. Research Progress in Woody Forage Silage [J]. Biotechnology Bulletin, 2021, 37(9): 48-57. |
| [5] | JIANG Fu-gui, CHENG Hai-jian, WEI Chen, ZHANG Zhao-kun, SU Wen-zheng, SHI Guang, SONG En-liang. Effects of Addition Amount of Molasses on the Fermentation Quality and Microbial Diversity of Hybrid Broussonetia papyrifera L. Vent Silage [J]. Biotechnology Bulletin, 2021, 37(9): 68-76. |
| [6] | DILIREBA·Abudourousuli , MUYESAIER·Aosiman , ZULIHUMAER·Rouzi , MA Qin, LEI Rui-feng, AN Deng-di. Advances on Microbial Diversity and Biological Improvement of Saline-alkali Soil [J]. Biotechnology Bulletin, 2021, 37(10): 225-233. |
| [7] | HUANG Ting, FANG Yuan, FENG Zhou, SHEN He, NIE Yong, ZHENG Xin, WANG Jia-quan, XU Zi-mu. Bacterial Communities in a Middle School Campus Assessed by High-throughput Sequencing [J]. Biotechnology Bulletin, 2020, 36(8): 96-103. |
| [8] | XIE Xian, LIANG Jun, ZHANG Ming, HU Rui-rui, CHENG Yuan, ZHANG Xing-yao. Endophytic Fungi Diversity in the Needles of Pinus densiflora with Sphaeropsis sapinea [J]. Biotechnology Bulletin, 2020, 36(2): 119-125. |
| [9] | KANG Jie, ZHANG Shu-yan, HAN Tao, SUN Zhi-mei. Microbial Diversity and Community Structure Characteristics of Yam Rhizosphere Soil at Different Development Periods [J]. Biotechnology Bulletin, 2019, 35(9): 99-106. |
| [10] | KANG Jie,ZHANG Shu-yan, HAN Tao,SUN Zhi-mei, LUO Tong-yang. Research on Rhizosphere Soil Microbial Diversity of Two Typical Kinds of Disease in Yam [J]. Biotechnology Bulletin, 2017, 33(7): 107-113. |
| [11] | GAO Xiu-zhi ,YI Xin-xin, LIU Hui ,WANG Xiao-dong ,CUI Zong-jun. Microbial Diversity of Traditional Soybean Paste During Fermentation in Northeastern China [J]. Biotechnology Bulletin, 2016, 32(4): 251-255. |
| [12] | Wu Yanyan, Qian Xixi, Li Laihao, Yang Xianqing, Ma Haixia. Research Progress on Diversity of Microbial Community During the Pickled Processing of Salted Fish Products [J]. Biotechnology Bulletin, 2015, 31(7): 40-44. |
| [13] | Feng Xuezhen, Wu Shanguang, Lu Yuan. Preliminary Application of PCR-DGGE to Analyzing Microbial Diversity of Ulva lactuca L. and Dictyota dichotoma [J]. Biotechnology Bulletin, 2014, 0(12): 73-77. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||